Abstract
Low-temperature stress (LTS) is one of the major abiotic stresses that affect crop growth and ultimately decrease grain yield. The development of rice varieties with low-temperature stress tolerance has been a severe challenge for rice breeders for a long time. The lack of consistency of the quantitative trait loci (QTLs) governing LTS tolerance for any given growth stage over different genetic backgrounds of mapping populations under different low-temperature stress conditions remains a crucial barrier for adopting marker-assisted selection (MAS). In this review, we discuss the ideal location and phenotyping for agromorphological and physiological parameters as indicators for LTS tolerance and also the traits associated with QTLs that were identified from biparental mapping populations and diverse rice accessions. We highlight the progress made in the fields of genome editing, genetic transformation, transcriptomics, and metabolomics to elucidate the molecular mechanisms of cold tolerance in rice. The stage-specific QTLs and candidate genes for LTS tolerance brought out valuable information toward identifying and improving LTS tolerance in rice varieties. We showed 578 QTLs and 38 functionally characterized genes involved in LTS tolerance. Among these, 29 QTLs were found to be colocalized at different growth stages of rice. The combination of stage-specific QTLs and genes from biparental mapping populations and genome-wide association studies provide potential information for developing LTS-tolerant rice varieties. The identified colocalized stage-specific LTS-tolerance QTLs will be useful for MAS and QTL pyramiding and for accelerating mapping and cloning of the possible candidate genes, revealing the underlying LTS-tolerance mechanisms in rice.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Low-temperature stress
- Physiological indicators
- Stage-specific QTLs and genes
- Breeding strategies
- Genetic transformation
1 Introduction
Rice (Oryza sativa L.) is an important cereal crop, being the staple food for more than half of the world’s population, providing 21% of global human per capita energy (Nalley et al. 2017). Approximately one tenth of Earth’s arable land is planted to rice, which is the primary source of food. The demand for this staple crop has put more pressure on rice breeders and biotechnologists to intensify rice production systems to enhance yield productivity under drastic changes in global climatic variations (GCVs). Based on the projection of global population growth, rice production must increase its annual yield by 1.2–1.5% in the coming decades to ensure global food security (Seck et al. 2012).
Rice is grown globally in diverse ecosystems, ranging from a few meters below sea level to as high as 2700 m above mean sea level (amsl). Despite rice originating in the swampy areas of the tropics, it is susceptible to a wide range of abiotic stresses (Ranawake and Nakamura 2011). Changes in GCVs have shifted the distribution of temperature variability across the globe. These remarkable shifts have resulted in more frequent low-temperature stress/cold stress events (chilling stress and freezing stress) during the rice-growing season, especially in subtropical and temperate regions, with consequent adverse effects on rice production. Low-temperature stress (LTS) is one of the major abiotic stresses that significantly decrease rice grain yield and is experienced by 10% of the total 130 million ha of rice (Mohanty et al. 2012). For instance, rice farmers have suffered significant declines in grain yield ranging from 0.5 to 2.5 t/ha in Australia, with an average yield income loss of USD 23.2 million/year because of LTS (Farrell et al. 2001).
Low temperature affects the rice industry in Africa, Asia, Australia, Europe, and South and North America. In the mountainous regions of South Korea, extremely low temperatures severely damaged rice crops in 1980 and 1993, with grain yield dropping by 26.0% and 9.2%, respectively, compared with the national average yield during those years (Schiller et al. 2001). Also, severe grain yield losses due to LTS conditions were reported in Italy, the United States (Board et al. 1980), and Chile. In India, LTS occurs in about 60% of the rice area in the northeastern and western hill states of the Himalayas, with cold stress caused by the cold irrigation water from melted snow and low ambient air temperature. LTS also directly affects crop duration, which increases relatively with cold temperature, thereby limiting to a large extent the possibility of double cropping in areas where water control is possible (Matlon et al. 1998).
Rice cultivars vary prominently in their tolerance of LTS, with subspecies indica more sensitive to LTS, while japonica cultivars are known to tolerate cold stress (Kim and Tai 2013). The rice crop is relatively sensitive to temperatures below 15 °C, which causes varying effects across different crop growth stages such as germination, seedling, vegetative, reproductive, and grain maturity (Andaya and Mackill 2003a, b). Low temperatures directly affect the crop by causing slow growth and decreased seedling vigor (Ali et al. 2006) as well as a delayed and lower percentage of germination (da Cruz and Milach 2000). At the seedling stage, manifestations of cold stress include low numbers of seedlings, decreased tillering, increased plant mortality, and induced nonuniform crop maturity (Zhang et al. 2014b). At the vegetative stage, LTS increases the growth period as exhibited by leaf discoloration or yellowing, leaf rolling or wilting, slowed growth, poor germination and seedling establishment, and the presence of rotten and dead seedlings (Lone et al. 2018). During flowering, the most sensitive stage, low temperature brings anomalies at anthesis, resulting in the cessation of anther development, nonripening of pollen, nonemergence of anthers from spikelets, improper anther dehiscence, pollen grains remaining in anther loculi, poor pollen shedding, and failure of pollen to germinate after reaching the stigmas (Suh et al. 2010; Shakiba et al. 2017).
LTS in both temperate and high-altitude rice-growing areas in the tropics and subtropics causes damaging effects throughout the growth dynamics of the crop (Ranawake and Nakamura 2015). The effect of LTS on different plant growth stages (germination, seedling, and reproductive) is crucial. In addition, there is a need for identifying an ideal location for phenotypic screening under LTS conditions, especially for agronomic, physiological, and biochemical traits to help in the development of LTS-tolerant cultivars. The establishment of genetic and genomic resources for LTS tolerance is a vital step toward the development of LTS-tolerant varieties. Over the years, the genetic and physiological perspectives of cold tolerance have been extensively studied, giving way to the development of a diverse set of criteria for evaluating the cold-tolerance phenomenon in rice at different growth stages. The rapid development of molecular markers and next-generation sequencing technology tools such as bisulfite sequencing and whole-transcriptome shotgun sequencing have been accelerated in many crop plants (Fig. 1). Several genomic regions have been studied for LTS in rice using biparental mapping populations and association mapping procedures. In this review, we have tried to organize and discuss the stage-specific QTLs and candidate genes for LTS tolerance, which could be used in LTS-tolerance rice varietal improvement programs. We also provide here the phenotypic characterization of LTS-tolerance traits at different growth stages of rice and associated genomic regions from the literature along with the traits. We also cover genome editing, genetic transformation, transcriptomics, and metabolomics tools for elucidating the molecular mechanisms of LTS tolerance in rice.
2 Phenological, Physiological, and Biochemical Indicators of LTS Tolerance at Different Developmental Stages
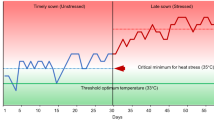
Screening for LTS in rice can be done at various growth stages (Table 1). In controlled conditions, LTS screening can be achieved timely with precision; however, it restricts the population in both sample size and number of samples. Thus, to screen large-sized populations, LTS breeding programs have resorted to evaluating many populations using cold water under field conditions (Snell et al. 2008). Such cold-water screening under field conditions has been established in research stations in Japan (Nagano 1998) and Korea (Lee 2001). The air temperature thresholds at the reproductive stage for cold-sensitive and cold-tolerant varieties are 20 °C and 15 °C, respectively (Satake 1976). Hence, high-elevation areas with low air and water temperatures, especially in subtropical regions in Kunming, People’s Republic of China (subsequently “China”), and in regions of Kashmir and Himachal Pradesh, India, are ideal spots for screening for cold tolerance (Jiang et al. 2012). Natural cold-screening hotspots that represent the target population of environments are vital for the systematic screening of germplasm and segregating breeding materials. The selection of such hotspots is crucial for the success of breeding and molecular genetic studies.
Rice is quite sensitive to LTS, mainly in tropical and subtropical regions at different growth stages. The critical temperature of the germination and reproductive stage at 15 and 17 °C has shown a significant impact on growth stage and yield decrease. However, the optimum temperature required for rice cultivation ranges from 25 to 35 °C (Yoshida 1981). The selection of LTS-tolerant rice varieties with a short duration is the key requirement for decreasing LTS damage. The effects of LTS in different growth stages, such as germination stage (GS), seedling stage (SS), and reproductive stage (RS), have significant impacts on agromorphological changes and yield component losses, especially in tropical zones. As compared to indica or indica × japonica backgrounds, japonica rice varieties have shown a wide range of LTS tolerance (da Cruz et al. 2013). The list of some LTS-tolerant rice varieties provided in Table 2 spans different countries, and most of these varieties are japonica type. However, some indica rice varieties also showed considerable LTS tolerance at the GS or SS (Biswas et al. 2017).
A few varieties have been proven to have a better performance for LTS in stage-specific growth conditions: for instance, Jinheung, Nipponbare, RNR 18805, and Italica Livorno for the GS (Miura et al. 2001; Fujino et al. 2004); M202, Lemont, and AAV002863 for the SS (Andaya and Mackill 2003b; Lou et al. 2007); and Norin PL8, Kirara397, RNR 17813, Akshaydhan, Taramati, WGL 44, Bhadrakali, JGL 3844, and WGL 44 for the RS (Saito et al. 2001; Kuroki et al. 2009). However, four rice varieties, B55, Banjiemang, Lijiangheig, and HSC55 from China and the United States, showed a consistent tolerance in three different growth stages (GS, SS, and RS) in rice (Basuchaudhuri 2014). For a further selection of LTS-tolerant rice varieties, several screening methods have been proposed, along with their pros and cons, for LTS-tolerant genotypes (Almeida et al. 2016). The selection of promising rice genotypes under natural LTS might favor negative results because of unpredictable climatic alterations in terms of stress intensity and duration of LTS. However, using high-throughput screening techniques such as image analysis, yield trait score, and robotics in controlled conditions of temperature, water, and air might help to detect tolerant genotypes and could also elucidate the traits related to morphological, biochemical, and yield-attributed traits during the plant growth period (Yang et al. 2014). Earlier studies of Snell et al. (2008), Suh et al. (2010), and Khatun et al. (2016) mentioned having developed reliable and straightforward screening methodologies for the selection of LTS-tolerant rice genotypes by preparing specific tanks for imposing cold-water irrigation and using a phytotron cabinet and low temperature in the glasshouse at different growth stages, which can provide the critical component traits. The primary focus traits for the GS related to germination rate, germination index, coefficient of germination, coleoptile length, and radicle length and also associated with early seedling vigor could be important traits for the selection of LTS tolerance at the GS (Li et al. 2018). In the SS, leaf discoloration, seedling survivability, leaf chlorophyll content, and estimation of the concentration of osmoprotectants (spermine and glycine betaine) and trehalose accumulation could be useful indicators to detect LTS at the SS (Han et al. 2004; Lou et al. 2007; Suh et al. 2012). Similarly, seed-setting rate, pollen growth development, incomplete panicle exsertion, days to flowering, spikelet fertility, and grain yield are the key traits for selection criteria at the RS (Ye et al. 2009; Jena et al. 2012; da Cruz et al. 2013). However, the natural incidence of LTS is significantly influenced to alter tolerance trait expression during phenotypic evaluation. Therefore, a combination of advanced molecular marker technology and high-throughput screening technologies provides the best method for prospecting for LTS-tolerant genotypes.
Several protocols exist to screen for cold tolerance/sensitivity in rice using different physiological and biochemical indicators. Two good indices of cold tolerance are seedling survival percentage (SSP) after subjecting seedlings to different low-temperature regimes (Morsy et al. 2007) and seedling chlorosis (Nagamine 1991). Many researchers have used SSP to analyze the resistance of transgenic plants to low temperatures (Chen et al. 2012; Huang et al. 2012). Nevertheless, the drawback of information obtained from SSP is that it is neither reproducible under natural conditions nor feasible for QTL studies. On the other hand, seedling chlorosis or the decrease in chlorophyll and leaf yellowing induced by cold stress could be captured by Soil Plant Analysis Development (SPAD) values to provide a more accurate measurement of cold stress at the seedling stage over a visual score. This indicator gives the direct association of the photosynthetic activity of the leaves, with low-temperature intensity and duration, as one of the yardsticks to screen rice germplasm and populations against cold stress (Hussain et al. 2018).
The accumulation of more dry matter and the functionality of photosystem-II (PSII) provide quantitative information on plant performance under cold-stress conditions (Gururani et al. 2015). An increase in the efficiency of PSII photochemistry gives information on the structural and functional changes in the photosystem of different plant types or transgenics, especially when the seedlings are exposed to low temperatures (Bonnecarrère et al. 2011). A sudden drop in chlorophyll integrity parameter and chlorophyll fluorescence (Fv/Fm) indicates a gas exchange decrease caused by alterations in the photosynthetic system. Therefore, combined information on gas exchange analysis and chlorophyll fluorescence is necessary to study the photosynthetic process (Saad et al. 2012) under cold stress. The expression of the AISAP gene of Aeluropus littoralis in rice confers broad tolerance of several abiotic stresses through the maintenance of photosynthetic apparatus integrity (Saad et al. 2012), particularly for PSII. The AISAP gene has become the tool to precisely evaluate for cold tolerance as it is related to final photosynthetic activity (da Cruz et al. 2013).
Biochemical parameters such as electrolyte leakage (EL), proline (Pro), and ascorbic acid (AA) were reported to be higher in sensitive variety IR50 than in resistant cultivar M202 (Kim and Tai 2011). Lee et al. (1993) showed that the exogenous application of abscisic acid (ABA) biosynthetic inhibitors resulted in low accumulation and low survival of seedlings under cold stress. Breeding varieties that accumulate higher concentrations of osmoprotectants (spermine and glycine betaine) was seen to be a strategy to overcome stresses (Yang et al. 1996), which has been proven through the development of transgenic rice that accumulates higher glycine betaine and shows resistance to LTS (Sakamoto and Murata 2002). Furthermore, the significant induction in the expression of antioxidative enzymes such as catalase (CAT), superoxide dismutase (SOD), and ascorbate peroxidase (APEX) under cold stress (Kuk et al. 2003) explained the rate of cold tolerance (Morsy et al. 2007) by RNA interference (RNAi) (Song et al. 2011) and in transgenic rice encoding Cu/Zn superoxide dismutase (sodC1) (Lee et al. 2009a, b).
The high tolerance of rice of cold stress could also be attributed to trehalose accumulation (Ge et al. 2008). Song et al. (2011) also found that accumulated amounts of trehalose can be used as an index for low-temperature tolerance/sensitivity. Increased amounts of trehalose through the overexpression of the OsNAC5 gene in transgenic rice plants were found to result in improved PSII function under abiotic stress conditions as it restricted damage due to photooxidation and exhibited soluble carbohydrates 20% higher than in nontransgenic plants (Garg et al. 2002). Similarly, at the reproductive stage, no sugar (sucrose and hexoses) accumulation has been found in anthers of low temperature-tolerant lines, resulting in no pollen grain sterility (Oliver et al. 2007). The overexpression of the gene OsAPXa (ascorbate peroxidase) in transgenic rice lines resulted in increased fertility under cold stress (Sato et al. 2011). It is also reported that unsaturated fatty acid content is related to plasma membrane stability at cold temperatures during the vegetative stage. Tolerant genotypes exhibited an increase in the amount of linolenic acid and a decrease in palmitic acid (da Cruz et al. 2010). Therefore, lipid peroxidation (Zhang et al. 2012a), along with EL (Huang et al. 2012), can be used to evaluate membrane lipid damage, which is an indirect assessment of cold tolerance.
Below the soil surface, roots play a crucial role under chilling stress, and root hydraulic conductivity (Lpr) is found to be profoundly affected when the plants are exposed to cold stress (Yamori et al. 2010). Murai-Hatano et al. (2008) found that Lpr decreased when susceptible rice genotypes were exposed to a temperature of 15 °C and the decrease was linked to transmembrane proteins, such as the aquaporins. These physiological and biochemical methods used for evaluating stress in rice genotypes and transgenic rice plants have played a significant role in understanding the crop’s mechanism of response against cold. However, most of these procedures are destructive, time-consuming, and stage-specific and are also inadequate and inappropriate for breeding programs involving the evaluation of many lines with large sample sizes. Therefore, to better understand cold-tolerance mechanisms, it is indispensable to study the phenomenon at the molecular level.
To improve the tolerance of rice of LTS, it is imperative to understand it at the molecular and physiological levels. At changing temperatures, rice plants modify their biological pathways, and molecular alterations occur within a different growth stage (Xiao et al. 2018; Ding et al. 2019). At different growth stages of rice plants, the initial effects of LTS are a decline in plasma membrane fluidity and transportation mechanism and alterations in physiological and metabolic activities, leading to disturbance of signaling processes (Ding et al. 2019). The cascades of the signaling process were followed by adjusting their cellular metabolism by activating the plasma membrane transporters and altering the metabolic responses (Fig. 2). These changes occurred in the intracellular levels by increasing abscisic acid concentrations via changes in growth hormones such as auxin and gibberellins and cross talk between the ethylene and salicylic acid signaling mechanism (Ghosh et al. 2016; Moraes De Freitas et al. 2016). These mechanisms have occurred through an alteration of membrane fluidity and the rearrangement of the cytoskeleton by the influx of calcium, which can trigger a downstream response to LTS tolerance by C-repeat binding factor: CBF-dependent (C-repeat/drought-responsive element-binding factor-dependent) and CBF-independent transcriptional pathways (Chinnusamy et al. 2010; Ma et al. 2015). Different growth stage-specific LTS-tolerance genes can be classified into three major groups as transcription factors, protein kinase genes, and functional genes, which may be involved in signal transduction pathways. Mainly, the CBF transcription factor regulates cold-responsive gene (COR) expression by binding to the CRT/DRE element. The promoter sequence of the CBF region is activated by the bHLH transcriptional activator of the inducer of CBF expression (ICE), which can also induce the expression of CBF genes toward LTS tolerance (Ito et al. 2006; Su et al. 2010). In addition to CBF pathway-related transcription factors, two genes, FRO1 (FROSTBITE 1) and OsFAD2, encode ferric reduction oxidase 1 and fatty acid desaturase 2, which are involved in LTS-tolerance mechanisms by maintaining membrane fluidity (Bevilacqua et al. 2015). The influx of calcium signals has also been associated with nitric oxide, reactive oxygen species, and mitogen-activated protein kinases, which can trigger the cascades of signaling pathways leading to LTS tolerance (Yuan et al. 2018). LTS tolerance at the germination stage is an important component trait for rapid seedling growth and uniform crop establishment, especially in the direct-seeding production system. The overexpression of the zeta class of glutathione S-transferases (OsGSTZ1) significantly improved germination rate and seedling growth under LTS (Takesawa et al. 2002). Similarly, Jin et al. (2018) identified a novel zinc finger transcription factor (OsCTZFP8) and it plays a key role in LTS tolerance at the reproductive stage in pollen fertility and seed setting along with yield per plant. Therefore, studying LTS-tolerance mechanisms at specific growth stages is crucial and may provide a better understating of key gene functions and their role in developing LTS-tolerant rice varieties in future breeding programs.
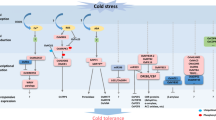
Sequential steps involved in the triggering of the signaling cascades for low-temperature stress (LTS) tolerance. Schematic representations of the LTS signal mostly processed by various biological processes such as stress perceptions and physiological and molecular responses. (1) LTS signaling initiated by ABA accumulation, and this is transduced to ABRE-containing NAC genes, which regulate the expression of NACRS genes for tolerance of LTS (Hu et al. 2008). (2) The higher concentration of ABA-induced pollen sterility occurs by increasing the expression of ABA biosynthetic genes OsZFP1 and OSNCED3 that convert zeaxanthin to xanthoxin. The LTS-tolerant plants were followed by ABA catabolism with a higher expression of two ABA-8-hydroxylase genes and further reduced to ABA concentration in anthers via C-8 hydroxylation pathways (Ji et al. 2011; Sharma and Nayyar 2016). (3) Increasing the influx of Ca2+ signals mediated by the DREB-CRT/DRE pathway under LTS, which is transduced by calcium-dependent protein kinases (CDPKs), and MYB family transcription factors induce the stress-responsive genes. OsMYB3R-2 regulates the LTS-tolerance mechanism at the seedling stage. OsMYB3R-2 may regulate through OsCPT1, which is involved in the DREB/CBF pathway in rice (Su et al. 2010). (4) MYBS3 is a single DNA-binding repeat MYB transcription factor, which mediates sugar signaling and also tolerance of the LTS signaling pathway. Interestingly, MYBS3 has a distinct tolerance mechanism with short- and long-term adaptation of LTS tolerance by repressing the DREB1/CBF pathway and late and slow response to LTS tolerance. (5) The cascades of mitogen-activated protein kinase consist of three components (MAPKKK, MAPKK, and MAPK) activated by an excess of reactive oxygen species under LTS. Kumar et al. (2008) found that MAP kinases 4 and 6 are strongly regulated by LTS and salt stress at the seedling stage. Cytosolic thioredoxin (OsTrx23) has a potential negative regulator for MAPKs’ activity. (6) LTS can also decrease the endogenous levels of bioactive gibberellic acid (GA) by the transcriptional repression of two bioactive GA synthesis genes (GA20ox3 and GA3ox1) (Sharma and Nayyar 2016). GAs had cross-talk with other hormones to regulate the stress-response mechanism. The signal cascades of GA interact with the receptor GID1 (GA INSENSITIVE DWARF1) and GRAS family protein DELLA involved in pollen development. The two TFs (ABI3 and ABI5) bind with DELLA complex proteins, which can promote the expression of SOMNUS involved as a negative regulator of seed germination (Serrano-Mislata et al. 2017; Li et al. 2018). The cross talk with auxin and jasmonic acid-biosynthetic genes plays a major role in favoring germination under LTS. The LTS signaling and regulations of the expression of TFs and gene responses are indicated by arrows. Each pathway relates to different traits under LTS. These traits are SC stomata closure, LP lipid peroxidation, MS membrane stability, RWC relative water content, PS pollen sterility, CC cell cycle, LG leaf growth, SG seedling growth, GM germination, PD pollen development
3 Genes/QTLs Underlying LTS in Rice Detected by Linkage Mapping and GWAS
For tolerance of LTS, information on the chromosomal location of QTLs and genes is limited in different growth stage-specific traits in rice. We carried out a comprehensive literature survey, including a Gramene database (http://archive.gramene.org) search, and aggregated 578 cold-specific QTLs associated with various growth stages, including germination, seedling, and booting or reproductive stages. Among these QTLs, 239 (41.3%) were mapped through genome-wide association studies (GWAS), while 339 (58.7%) QTLs were identified from different types of biparental mapping populations, and detailed information is provided in Table 3. Based on the distribution of the reported QTLs on the chromosomes, the highest number of QTLs was noticed on chromosome 1 (65), followed by chromosome 7 (60), whereas the lowest number of QTLs was noticed on chromosome 8 (35) (Fig. 3a). Furthermore, based on the association of these QTLs with growth, stage-specific traits were classified into 214 QTLs related to GS, 249 QTLs for SS, and 115 QTLs for RS (Fig. 3b).
The physical positions of these stage-specific QTLs are depicted in Fig. 4. The QTLs were classified as main-effect QTLs (M-QTLs), based on the phenotypic variance explained by each QTL, which was ≥30%. Notably, five M-QTLs for GS-related traits (including germination rate, germination percentage, and germination index) were found on chromosomes 2, 5, and 7 (Xu et al. 2008; Cui et al. 2018); 15 M-QTLs for SS related to shoot and root growth traits on chromosomes 4, 6, 7, 8, 11, and 12 (Andaya and Mackill 2003b; Wang et al. 2009; Ranawake et al. 2014; Yu et al. 2018); and 13 M-QTLs for RS related to heading time, panicle weight, spikelet fertility, and culm length on chromosomes 1, 2, 4, 7, and 10 (Dai et al. 2004; Kuroki et al. 2009; Wainaina et al. 2018) were identified. A total of 29 QTLs were colocalized on all 12 chromosomes except on chromosomes 5 and 12 (Fig. 4 and Table 4). However, the colocalized stage-specific QTLs range from two to seven. Interestingly, the highest numbers of GS- and SS-specific QTLs were colocalized in the 22.5 Mb genomic region on chromosome 7. Three combinations of stage-specific QTLs (GS, SS, and RS) were identified on chromosome 10 in the 19.1 Mb region (Liu et al. 2013; Pan et al. 2015; Schläppi et al. 2017).
Diagram of LTS-tolerance QTLs by comprehensive literature survey and Gramene database (http://archive.gramene.org) in rice. On the right side, the numerical values in chromosome bars indicate the position of QTLs and genes in Mb, and on the left side, those indicate the QTLs for stage-specific and LTS-tolerance genes. A colored font represents the stage-specific QTLs (green: germination; blue: seedling; and red: reproductive/booting stage) and genes for LTS tolerance (pink). The octagonal shapes and numerical values represent the two different stage-specific QTLs that were colocalized in the same genetic region, and the square shape indicates the QTLs and reported LTS-tolerance genes aligned together in the same genetic regions of chromosomes
3.1 Germination Stage
Seed germination is of paramount interest for breeding varieties suitable for temperate regions and high-elevation areas, but it is given a lower priority than traits such as high yield and grain quality. To date, more than 200 QTLs (98 QTLs were identified from biparental mapping populations and 116 QTLs from GWAS) have been mapped on the 12 chromosomes (Fig. 3). The phenotypic variance of these QTLs ranges from 3.58% to 42.29%. The M-QTLs (≥30% PVE) for the GS were identified on chromosome 2 (qLTG-2-1), chromosome 5 (qLTG-5-2.1 and qLTG-5-2.2), and chromosome 7 (qGV7-1.1 and qGI7-1.2) (Xu et al. 2008; Yang et al. 2018). With a comprehensive analysis of GS-QTLs, nine genetic regions on eight chromosomes (Ch3: 17.2–17.8 Mb, Ch5: 21.5–21.6 Mb, Ch6: 5.4–6.2 Mb, Ch7: 1.7–2.7 and 20.13–22.6 Mb, Ch9: 21.9–24.6 Mb, Ch10: 11.6–14.2 Mb, Ch11: 23.0–24.2 Mb, and Ch12: 7.0–7.1 Mb) had more than four GS-QTLs that were colocalized. Recently, Yang et al. (2018) identified 12 and 23 QTLs for low-temperature germinability (LTG) and cold tolerance at the seedling stage by using recombinant inbred lines (RILs) that were derived from a backcross population of Dongnong422 and Kongyu131. Interestingly, seven QTLs on chromosome 12 in the 7.1 Mb region and four QTLs on chromosome 7 in the 22.55 Mb region were colocalized, and they were associated with several GS traits such as germination time and rate, mean length of incubation time, coefficient of germination, germination value, mean daily germination, and germination index. Cloning and characterization of the major QTL qLTG3-1, conferring more than 30% of the variation (Fujino et al. 2004), revealed that this gene encodes for a protein of unknown function. At the same time, a microarray analysis indicated that a complex metabolic and signal pathway was involved (Fujino and Matsuda 2010). Genome-wide expression analysis suggested that genes involved in defense responses were upregulated by qLTG3-1 and played a more general role in germination (Fujino et al. 2008), whereas correlation with proteomics indicated its involvement in rice growth and adaptability (Fujino and Sekiguchi 2011). With genome-wide association mapping studies, Pan et al. (2015) identified significant 17 QTLs from the 174 mini-core collections of Chinese rice accessions, 45 QTLs from the 202 Rice Mini-Core Collections (Schläppi et al. 2017), and 54 QTLs from a global collection of 400 Rice Diversity Panel 1 (Shakiba et al. 2017) that were detected under low-temperature germinability in rice. Given these challenges and potential in the form of effective QTLs (E-QTLs) with colocalization of QTLs, the development of molecular markers for selection for LTG would significantly contribute to identifying and developing LTS-tolerant varieties.
3.2 Seedling Stage
Low-temperature stress severely affects the SS, causes slow growth, yellowing symptoms on leaves, drying of leaves, and decreased early seedling vigor, and ultimately leads to seedling death (Wang et al. 2011; Lone et al. 2018). Tolerance of LTS at the SS is one of the key stages to ensure stable early seedling growth in temperate and high-altitude regions. For the genetics of the SS, several researchers have identified SS-specific QTLs using different genetic backgrounds of mapping populations such as RILs, near-isogenic lines (NILs), introgression lines, doubled haploids, and segregating F2 and F3 families (Biswas et al. 2017; Liang et al. 2018; Sun et al. 2019). However, the genetic backgrounds of japonica donors as LTS-tolerant cultivars have revealed that several QTLs and genes are controlling LTS tolerance during the SS in rice. So far, more than 249 QTLs (both major and minor) have been mapped on all of the rice chromosomes responsible for LTS tolerance at the seedling stage (Table 3). Among the total QTLs, 159 were identified from biparental mapping populations and 90 from GWAS. The phenotypic variance of these QTLs ranges from 4.51% to 60.96%. Fifteen M-QTLs (≥30% PVE) were identified on six different chromosomes: 4, 6, 7, 8, 11, and 12. However, only a few of them were M-QTLs that were colocalized with different stage-specific QTLs. For instance, qSCT4.2 (57.62% PVE) on chromosome 4 (22.3–26.91 Mb) was shared with four QTLs (qRL-4-2_23d, q9d-4, qLTG-4-2, and qHD-4) for root length, percentage of plumule growth, germination rate, and heading date under LTS conditions (Miura et al. 2001; Wainaina et al. 2018; Yu et al. 2018). Similarly, qSCT8 (60.96% PVE) on chromosome 8 (24.6–27.8 Mb) was shared with cold-tolerance seedling-stage QTL qCTS8-1 (Andaya and Mackill 2003b), seed weight per plant QTL qSWTCT8-4 (Shakiba et al. 2017), and seedling survival rate QTL qLTSSvR8-2 (Pan et al. 2015). Importantly, two M-QTLs (qCTS12a and qCTS12b) for cold-induced wilting tolerance and cold-induced necrosis tolerance on chromosome 12 had PVE of 40.6% and 41.7% (Andaya and Mackill 2003b), respectively. The same genetic region of 7.1–9.3 Mb overlapped with ten QTLs for GS- and RS-related traits such as germination index, incubation time, coefficient of germination, and seed-setting rate (Pan et al. 2015; Yang et al. 2018). Other two M-QTLs (qGAS12 and qSCT12.1) had PVE of 42.9% and 53.09%, respectively. These QTLs were associated with the growth ability of seedlings at low temperatures (Han et al. 2004) and seedling cold tolerance (Yu et al. 2018) on chromosome 12. The genetic region of 24.2–25.0 Mb was shared with GS- and SS-specific QTL qCTS12.1 (Wang et al. 2011), qLTSS12-2 (Schläppi et al. 2017), and qSWTCT12 (Shakiba et al. 2017).
The seedling-stage QTLs (seven genetic regions on seven chromosomes) were found to overlap with more than five SS-specific QTLs. The highest number of QTLs was found to overlap in the 16.3–18.4 Mb region on chromosome 10 (12 QTLs), followed by 11 QTLs on chromosomes 2 and 11 (at 14.3–17.7 and 6.8–11.3 Mb), six QTLs on chromosome 3 (2.7–3.7 Mb) and chromosome 9 (1.1–4.8 Mb), and five QTLs on chromosome 7 (22.5–23.8 Mb) and chromosome 11 (24.9–25.4 Mb), respectively. However, two chromosomes (7 and 11) have significant M-QTLs that are also colocalized with more than five QTLs. In a deeper understanding of these two chromosomes, the M-QTL qCTS7(2) on chromosome 7 (20.0–20.9 Mb) had PVE of 35.3%, and it overlapped with two other QTLs (qCTGERM7-4 and qCTB-1.7) related to GS and RS (Dai et al. 2004; Ranawake et al. 2014; Shakiba et al. 2017). Another M-QTL, qCTS11(1)-2 on chromosome 11 (20.5–26.0 Mb), had PVE of 35.6% (Ranawake et al. 2014), the same region was associated with a seed germination recovery rate QTL (qGRR11) (Jiang et al. 2017), and two SS-specific QTLs (qCTS11-9 and qCTS11-10) were identified from the GWAS analysis (Wang et al. 2016). Therefore, the combination of stage-specific QTLs with GS, SS, and RS in the same genetic region could be a promising site for identifying potential candidate genes for improving LTS tolerance in the seedling stage.
3.3 Booting/Flowering Stage
Unlike the GS and SS, the RS is highly sensitive to LTS. Many traits, such as microspore abortion, no anther dehiscence, high spikelet sterility, incomplete panicle exsertion, delayed heading, and failure to produce pollen grains, are affected by LTS in the RS (da Cruz et al. 2013; Liang et al. 2018). Several studies have detected and mapped many major- and minor-effect QTLs responsible for LTS tolerance at the booting/flowering stage using different genetic backgrounds of mapping populations (Table 3). So far, more than 100 QTLs have been mapped on all of the rice chromosomes. The QTLs for tolerance at the RS showed that 81 QTLs from biparental mapping populations and 33 QTLs from GWAS were identified from the comprehensive literature survey. The phenotypic variance of these QTLs ranges from 2.94% to 71.0%. A total of 14 M-QTLs (≥30% PVE) were located on six different chromosomes (1, 2, 4, 7, 8, and 10). Saito et al. (1995) reported two QTLs on chromosomes 3 and 4 responsible for cold tolerance at the booting stage from Norin PL8. Saito et al. (2001) also reported two OTLs (Ctb-1 and Ctb-2) on chromosome 4 governing spikelet fertility under cold stress by using a set of NILs derived from a cross between cold-tolerant rice variety Norin PL8 and cold-sensitive commercial variety Kirara397 from northern Japan. By using cool-water irrigation (19 °C), Kuroki et al. (2009) identified five QTLs for cold tolerance at the booting stage, four QTLs for days to heading, and three QTLs for culm length on chromosomes 1, 2, and 10. One of the major QTLs (qCTB.1) flanked by RM1003 and RM3482 on chromosome 1 associated with cold tolerance at the booting stage was discovered after 3 years of field trials at the National Agricultural Research Centre for Hokkaido Region, Sapporo, Japan. Similarly, eight QTLs for the booting stage were identified on chromosomes 1, 4, 5, 10, and 11 by using a set of NILs from a cold-tolerant japonica landrace (Kunmingxiaobaigu) and cold-sensitive japonica cultivar (Towada) (Xu et al. 2008). However, four QTLs (qCTB-1-1, qCTB-4-1, qCTB-5-1, and qCTB-5-2) were detected in two different environments.
A total of nine genetic regions on chromosome 1 (28.6 and 36.6 Mb), chromosome 2 (21.5 Mb), chromosome 4 (24.5 Mb), chromosome 7 (8.1 and 18.1 Mb), chromosome 8 (11.5 Mb), and chromosome 10 (9.2–9.6 and 20.0–25.2 Mb) were associated with tolerance of RS traits such as heading time, booting stage, culm length, spikelet fertility, spikelet number, and panicle weight (Dai et al. 2004; Kuroki et al. 2009; Wainaina et al. 2018). The M-QTL (qCTB_1) on chromosome 1 for cold tolerance at the booting stage had PVE of 47.3% (Kuroki et al. 2009), and it was close to the genetic region associated with SS-specific QTL qCTSR1-3 (36.9 Mb) (Zhang et al. 2018) and MYB transcription factor OsMYB3R-2 (36.1 Mb), which is bound to the mitotic-specific activator during LTS tolerance (Ma et al. 2009). The overexpression of this gene significantly enhances the many transcripts for G2/M phase-specific genes in response to cold stress. Another M-QTL (qSNP_1; 28.6 Mb) in the genetic region on the same chromosome overlapped with Osa-MIR319a. In rice, the miR319 gene family comprises two genes, Osa-MIR319a and Osa-MIR319b. Both of them are significantly involved in increasing leaf blade width under cold tolerance (Yang et al. 2013a). The colocalization of stage-specific QTLs on chromosome 2 is associated with nine QTLs (21.1–21.8 Mb) for GS- and SS-specific QTLs (Lou et al. 2007; Shakiba et al. 2017; Yang et al. 2018). Similarly, the M-QTL qPW-7 (PVE of 41%) on chromosome 7 overlapped with cold-tolerance seedling-stage QTL qCTS7-3 (Wang et al. 2016). Six M-QTLs were associated with tolerance of RS traits on chromosome 10. In the interval regions of M-QTLs, two QTLs (qLTG10-2, 21.1 Mb; qCTGERM10-4, 22.3 Mb) and one DNA-binding repeat MYB transcription factor (OsMYBS3, 22.1 Mb) are responsible for cold tolerance at the GS and SS (Su et al. 2010; Schläppi et al. 2017; Shakiba et al. 2017). The overexpression of MYBS3 confers chilling tolerance, and it has also been associated with MYBS3-mediated cold signaling pathways (Su et al. 2010). Taken together, the stage-specific QTLs of different combinations of GS, SS, and RS and associated candidate gene results could prove to be useful in breeding programs for low temperature-tolerant rice lines. These M-QTLs along with colocalized stage-specific QTLs have great potential for use in the future as their application through marker-assisted selection will hasten the process of developing cold-tolerant rice varieties for temperate and high-altitude ecosystems.
The LTS-tolerance QTLs mentioned above included a comparison of stage-specific QTL positions across different genetic backgrounds of mapping population studies. Such comparisons of stage-specific QTL positions are more informative. These QTLs may harbor potential candidate genes related to LTS, thus providing valuable information to develop LTS-tolerant rice cultivars. For this, the functionally characterized LTS-tolerance genes were collected from the OGRO database (Overview of Functionally Characterized Genes in Rice Online database) on the Q-TARO website (http://qtaro.abr.affrc.go.jp/ogro). A total of 38 candidate genes were involved in LTS tolerance in different parts of the rice plant. Specific gene functions for LTS tolerance and stages are mentioned in Table 5 and are also mapped on the genetic map (Fig. 4). Among these candidate genes, the majority of them were associated with the SS (25 genes), followed by two genes for the RS. The remaining eight genes were for the SS and RS, two genes for the GS and SS, and a single gene for the GS and RS. Functionally, the candidate genes were involved in altering the various metabolic and physiological pathways in different growth stages of LTS-tolerance mechanisms. Seven genetic regions on four chromosomes (17.9 Mb on chromosome 1; 25.6 and 34.4 Mb on chromosome 3; 15 and 25.7 Mb on chromosome 5; and 23.9 and 26.3 Mb on chromosome 6) are colocalized with candidate genes and QTLs. For example, OsSPX1 is colocalized with qCTS6-5 at 23.9 Mb on chromosome 6. A previous study revealed that OsSPX1 plays a key role in the cross talk between cold tolerance, phosphate homeostasis, and oxidative stress tolerance in the SS (Wang et al. 2013a), and the cold tolerance QTL (qCTS6-5) was mapped in the SS by using GWAS (Wang et al. 2016). Based on the fine-tuning of the interval regions of the M-QTLs, several researchers have identified many candidate genes for LTS tolerance. The M-QTLs and colocalized QTLs within identified genetic regions, especially on chromosomes 1, 3, 5, 7, and 10, may be potential genomic regions to introgress into existing moderately LTS-tolerant genotypes or mega-varieties to improve their rate of tolerance in marker-assisted breeding programs.
4 Molecular Mechanisms of LTS Tolerance
Several genes and transcription factors are involved in regulating the molecular pathways related to alteration of physiological and metabolic compounds and, further, reprogramming of their gene expression patterns against cold-stress tolerance mechanisms (Fig. 2). During LTS response, multiple sensors and signaling elements on the plasma membrane trigger the expression of COR (cold-responsive) genes via increasing cytosolic Ca2+ levels. This increase in Ca2+ is mediated by the ligand-activated Ca2+ channels. Further higher levels of Ca2+ in the cytosol lead to signal amplification through phospholipids (Williams et al. 2005; Hashimoto and Komatsu 2007; Chinnusamy et al. 2010), which are sensed by calcium-binding proteins and other transcription factors regulating the expression of LTS-tolerance genes, which can ultimately lead to adaptation and survival during cold-stress conditions (Shinozaki and Yamaguchi-Shinozaki 2000). However, the changes in various gene expression patterns are governed by a signal cascade mechanism, which also triggers the formation of plant hormones (abscisic acid, salicylic acid, and ethylene) that may be involved in integrating various stress signal pathways and controlling downstream stress responses. CBF (C-repeat/DREB [drought-responsive element-binding factor]) regulon is a highly conserved cold-response pathway (Chinnusamy et al. 2010). The ICE1–CBF transcriptional cascade plays a crucial role in cold acclimation (Zhang et al. 2004). Constitutively expressed ICE1 (inducer of CBF Expression 1) binds to the CBF promoter to activate cold-resistance genes, and overexpressing ICE1 has significantly enhanced cold tolerance in Arabidopsis (Chinnusamy et al. 2003). Similarly, ICE2 (At1g12860, a homolog of ICE1) overexpression demonstrated enhanced freezing tolerance in Arabidopsis after cold acclimation (Fursova et al. 2009). The report published showed that it interacts with the alpha subunit of the sole heterotrimeric G protein, leading to a cytosolic Ca2+ signal or itself behaving as a cold-sensing calcium channel. A Ca2+ signal may be mediated by CPKs and CBL-CIPKs, which in turn activate MAP kinases (Yang et al. 2010). Phosphorylation of transcription factors such as that of CAMTAs and ICE1/2 is supposed to be caused by activated MPKs, which in turn activate COR genes (Zhu 2016).
Most COR genes carry C-repeats or DREBs (CCGAC cis-element) in their promoters, which bind to CBFs to activate their expression (Chinnusamy et al. 2007). The induction of CBF1, CBF2, and CBF3 precedes the COR genes in response to cold stress. Constitutive overexpression studies in Arabidopsis revealed redundant functional activities of CBF1, CBF2, and CBF3 and showed different functions in cold acclimation (Gilmour et al. 2004). This was observed in the cbf2 T-DNA insertion mutant with enhanced tolerance of freezing (with or without cold acclimation), dehydration, and salt stress through increased expression of CBF1 and CBF3. Furthermore, the cold-induced expression of CBF1 and CBF3 precedes that of CBF2, revealing a temporal difference in CBF expression. These results indicate that CBF1 negatively regulates CBF3 and CBF2 to optimize the expression of downstream target genes (Doherty et al. 2009). Transgenic analysis of CBF1 and CBF3 RNAi lines revealed that both CBF1 and CBF3 are required for the full set of CBF regulon expression and freezing tolerance (Novillo et al. 2004). While responding to cold stress, ICE1 and calmodulin-binding transcription activators (CAMTAs) bind to CBF3 and CBF2 promoters, respectively, to respond to their expression (Doherty et al. 2009). Furthermore, in many cellular signaling pathways, particularly in response to cold stress, protein phosphorylation is considered crucial, and it predicts the involvement of one or more protein kinases to phosphorylate ICE1 to help in CBF expression (Chinnusamy et al. 2007; Yang et al. 2010).
4.1 Signaling Pathways Leading to LTS Tolerance from the Cloned Genes
Transgenic and gene expression analysis has helped to understand the physiological mechanisms responsible for tolerance against various abiotic stresses, including LTS, in plants (Gao et al. 2008; Moraes De Freitas et al. 2016). Using the OGRO database on the Q-TARO website, we collected 38 candidate genes that have been functionally characterized for LTS tolerance in different stages of the rice plant (Table 5 and Fig. 4). Among the total number of genes, eight were involved in the two stage-specific tolerance mechanisms of LTS. Four genes on chromosome 1 (OsCOIN, 0.2 Mb; OsGSK1, 5.7 Mb; OsGH3-2, 32.2 Mb; and OsMYB3R-2, 36.1 Mb), two genes on chromosome 6 (OsiSAP8, 24.4 Mb; and OsbZIP52, 27.3 Mb), and a single gene on chromosome 4 (OsCAF1B, 34.9 Mb) and chromosome 11 (OsAsr1, 3.2 Mb) were associated with tolerance at the SS and RS in rice.
The promising genetic regions of OsGH3-2 (Greco et al. 2012) and OsMYB3R-2 (Ma et al. 2009) showed clear evidence of seedling survival rate and seed-setting rate under cold stress. The overexpression of OsGH3-2 significantly modulates abscisic acid (ABA) and endogenous indole-2-acetic acid (IAA) homeostasis, resulting in increased cold tolerance. Furthermore, two genes (OsTPP1 and OsFAD2) on chromosome 2 and a single gene (OsZFP182) on chromosome 3 were associated with the GS, SS, and RS. Expression analysis of OsTPP1 confers a tolerance mechanism for salt and cold by activating the transcriptional regulation pathways (Ge et al. 2008). The expression pattern of OsFAD2 under LTS in different tissues in young seeds, stems, roots, and leaves plays a significant role in membrane lipid desaturation and maintenance of the lipid balance in different photosynthetic tissue (Shi et al. 2012). Meanwhile, overexpression of OsZFP182 in transgenic lines showed an increasing accumulation of various osmolytes, which resulted in an increase in tolerance of drought, cold, and salt (Huang et al. 2012).
On chromosomes 5 and 10, two genes (OsLti6b and OsPRP3) are associated with the RS. OsLti6a and OsLti6b encode membrane proteins that contribute greatly to membrane stability (Morsy et al. 2005; Kim et al. 2007). OsPRP3 is a novel flower-specific prorich protein (PRP) that is significantly overexpressed in the RS under LTS, mainly in flower development (Gothandam et al. 2010). The remaining 25 candidate genes are involved in LTS tolerance in the SS. The important upregulated or overexpressed genes/TFs concerning their expression and function in different LTS stages and in cold stress conditions are described briefly in Table 5. The overexpression of several TFs and protein kinases, such as OsISAP8, OsbHLH1, OsDREB1/CBF, ROS-bZIP, SNAC2, OsCIPK12, OsNAC6, OsCOIN, OsMAPK5, OsMYB4, and OsISAP1, confers LTS tolerance in the SS in rice (Mukhopadhyay et al. 2004; Nakashima et al. 2007; Xiang et al. 2007; Kanneganti and Gupta 2008).
For tolerance in the RS, two cell wall acid invertase genes (OsINV1 and OsINV4) and one vacuolar acid invertase gene (OsINV2) were associated with low temperature at the pollen developmental stage. Among these genes, OsINV4 is anther-specific and is downregulated by cold treatment, consequently causing a disturbance in hexose production and starch formation in the pollen grains in the tapetum cells. However, no decrease in expression of OsINV4 vis-à-vis any sucrose accumulation in the anthers and pollen grains in a cold-tolerant cultivar (R31) was observed after cold treatment (Oliver et al. 2005). The OsMAPK5 gene codes for a protein involved in kinase activity usually induced by ABA and various biotic and abiotic stresses. OX lines for the OsMAPK5 gene exhibited increased tolerance of cold and other stresses (Xiong and Yang 2003). Thus, LTS tolerance at specific growth stages involved essential stress-responsive genes and TFs, which may be potential targets in genetic improvement for LTS tolerance in rice. However, the constitutive overexpression of these genes has led to metabolic instability and yield penalty and, as observed in so many experiments, has retarded growth under normal conditions, as shown by transgenic plants (Gilmour et al. 2000; Ito et al. 2006; Nakashima et al. 2007). Using stress-inducible promoters such as the rd29A promoter instead of constitutive promoters minimizes these side effects on plant growth (Kasuga et al. 2004). However, although the complex nature of the cold-tolerance phenomenon has been explained by transgenic technology, as many genes/TFs have been exploited and manipulated, its field utility is yet to be explored and assessed.
The important COLD1 gene of Nipponbare in the background of 93-11 exhibited tolerance in the rice SS by encoding a GTPase-accelerating binding factor that regulates G-protein signaling by sensing cold to trigger Ca2+ signaling for cold tolerance (Ma et al. 2015). Five QTLs (on chromosomes 1, 2, 4, 6, and 8) were reported from the cross between chilling-tolerant Nipponbare (japonica) and chilling-sensitive 93-11 (indica) cultivars at 4 °C of cold treatment. Among these QTLs, three (COLD2, COLD4, and COLD5) were found genetically interacting with each other, and together, they contributed PV of 16.8%, while COLD1 alone exhibited PV of 7.23%. Fine-mapping of COLD1 leads to the identification of sequential alterations at the first exon (SNP1) and fourth exon (SNP2) and five substitutions in introns (SNP3) in the 4.78-kb region of the COLD1j gene (LOC_Os04 g51180). The transgenic approach proved that the SNP2 allele (COLD1jap) had a significant overexpression compared to WT plants and suggested that COLD1 modulates chilling tolerance in rice. Therefore, a good combination of stage-specific QTLs and cold-tolerance genes could be helpful for developing LTS tolerance in rice. Despite the detection of cold-tolerance genes and QTLs, to date, none of the LTS-tolerant varieties were ever developed through marker-assisted backcrossing (MAB). This raises two questions: first, are the discovered LTS tolerance-related QTLs and genes verifiably useful for MAB? Second, is there a lack of training of LTS-tolerance rice breeders to exploit the advances in molecular genetics of LTS tolerance? However, a more reliable and reproducible QTL must be first identified to improve LTS tolerance through more breeder-friendly MAB approaches.
4.2 Genome-Wide Association Studies for LTS Tolerance
The recent development in high-throughput genome sequencing platforms, GWAS, has become a powerful tool to exploit linkage disequilibrium to dissect traits and identify the genomic regions associated with a trait of interest. GWAS have been used in various research efforts such as drought, salinity, and deficiency and toxicity tolerance to understand the trait associated with the whole-genome sequence of genotypes using a diverse set of rice germplasm accessions. The sequencing of rice genotypes is commonly classified into SNP array genotypes and resequenced SNP genotypes. Zhang et al. (2018) identified high-quality filtered reads with a call rate of 95% for 3867 SNP markers by genotyping 249 indica rice varieties using a 5K SNP rice array for cold tolerance at the bud burst stage. GWAS for severity of damage (SD) and seed survival rate (SR) revealed 47 SNP loci significantly associated with SD and SR in cold treatment at 5 °C for 5 days. Among these SNPs, the major QTL qCTSR1-2 on chromosome 1 overlapped with qCTSD1-2, which explains 13.2% of the total phenotypic variation. GWAS for germination and reproductive stages that Shakiba et al. (2017) conducted with the Rice Diversity Panel 1 (RDP1), which consisted of 400 O. sativa accessions belonging to five major subpopulations, resulted in the identification of 42 loci associated with cold tolerance, and several QTLs were colocalized with previously reported LTS-tolerance QTLs. Recently, Xiao et al. (2018) identified a potential candidate locus (LOC_Os10g34840) on chromosome 10, which is responsible for cold tolerance at the seedling stage, by assessing the total diversity panel of 1033 rice accessions with 289,231 SNP markers. The loci at 18.58–18.65 Mb overlap with previously reported cold-tolerance QTLs (Xiao et al. 2015), and, furthermore, they have been fine-mapped and validated by quantitative expression analysis. Similarly, using specific locus amplified fragment sequencing (SLAF-seq) technology, Song et al. (2018) conducted GWAS with 150 accessions of rice landraces by using high-density SNPs A total of 26 significant SNPs were associated with cold tolerance at the seedling stage. These SNPs had PVE ranging from 26% to 33%, and among them, three QTLs were colocalized with previously cloned genes such as OsFAD2, OsMYB2, and OsCIPK03 related to LTS tolerance at the rice seedling stage (Yang et al. 2012). Interestingly, Song et al. (2018) noticed a strong signal of trait-marker association peaks on chromosome 1, with PVE of 27%. The expression profiling and bioinformatics analyses reveal that a novel candidate gene (Os01g0620100) showed a significant difference between the cultivars tolerant and sensitive to LTS because of the polymorphism in the WD40 domain. Thus, Os01g0620100 is an important source for developing LTS tolerance by using marker-assisted selection. A comprehensive literature survey of GWAS for LTS tolerance resulted in a total of 239 QTLs distributed on all 12 chromosomes (Fig. 3). Among these, 116 QTLs for GS, 90 QTLs for SS, and 33 QTLs for RS were reported by several researchers (Pan et al. 2015; Wang et al. 2016; Sales et al. 2017; Schläppi et al. 2017; Shakiba et al. 2017; Singh et al. 2017; Zhang et al. 2018). The highest number of QTLs was detected on chromosome 2 (42), whereas the lowest number was detected on two chromosomes, 10 and 12 (14 QTLs). The physical position of each stage-specific QTL from GWAS revealed that three major genetic regions on chromosome 1 (41.39–41.86 Mb), chromosome 3 (3.03–3.76 Mb), and chromosome 6 (6.01–6.80 Mb) were colocalized with more than four QTLs. Two QTLs (qCTGERM1-8 and qSWTCT1-2) for GS and two other QTLs (qCTS1-4 and qCTS1-5) for SS stage-specific QTLs overlapped on chromosome 1 (Wang et al. 2016; Shakiba et al. 2017). Similarly, eight QTLs (qCTS3-6, qCTS3-7, qCTSR3-1, qCTSD3-1, qCTS3-8, qLTRSSR3-1, qLTSSR3-1, and qCTS3-9) detected on chromosome 3 for GS, SS, and RS were colocalized (Pan et al. 2015; Wang et al. 2016; Zhang et al. 2018). Four QTLs associated with three traits related to germination rate, cold tolerance at the seedling stage, and plumule recovery growth after cold exposure were colocalized on chromosome 6 for the GS and SS (Wang et al. 2016; Schläppi et al. 2017; Shakiba et al. 2017). Interestingly, five chromosomal regions at 30.1 Mb (chromosome 4), 5.1 Mb (chromosome 6), 28.3 Mb (chromosome 7), 14.2 Mb (chromosome 8), and 27.3 Mb (chromosome 12) were associated with the GS, and some RS-specific QTLs were also aligned together in the same genetic regions. With a large number of QTLs for stage-specific traits from GWAS information from the genome sequencing data, high-throughput phenotyping and various statistical methods could provide beneficial information for MAB programs and the discovery of potentially useful chilling-tolerance genes/alleles . Furthermore, the combination of gene expression profiling and omics technologies such as proteomics, metabolomics, epigenetics, and genome editing tools will facilitate the confirmation of more candidate gene functions in rice.
4.3 Transcriptomics Related to LTS Tolerance
Transcriptome sequencing has increased the accessibility of genomic resources in various crops, including rice. Transcriptome analysis using microarray technologies is one of the most powerful techniques that link sequence information directly to functional genomics (Sinha et al. 2018) and immensely contributes to understanding the specific tissue- or stress response-related genes in the molecular mechanisms of biotic and abiotic stress tolerance. Comparative transcriptome analysis provides a way to distinguish different genes that are regulated in stress tolerance in comparison to expression patterns between the homologous genes in various crops (Lee et al. 2019). In response to LTS tolerance, Yang and Poovaiah (2003) observed that, at low temperatures, commonly upregulated genes were associated with Ca2+ signal transduction. Several genes such as Ca2+-dependent protein kinases, calmodulin, mitogen-activated protein kinase 1, Ca-transporting ATPases, protein phosphatase 2C family proteins, and serine/threonine-protein kinases related to signal transduction pathways were identified in the endoplasmic reticulum-phase of chilling-stress tolerance (Yang et al. 2012; Zhang et al. 2012b). A key initial event that occurs in cold-stress response is the induction of the AP2/EREBP TF family, which includes CBF/DREB TFs, which are commonly induced within 30 min of cold treatment in plants (Zhang et al. 2004; Ito et al. 2006). CBF/DREB genes have also been shown to be gated by a circadian clock and to display cyclic behavior during cold stress (Gilmour et al. 2004). Previously, Bai et al. (2015), working on the anther transcriptome of photo-thermosensitive genic male sterile rice lines Y58S and P64S under cold stress, identified some differentially expressed genes (DEGs) involved in signal transduction, metabolism, transport, and transcriptional regulation. Among these DEGs, more differentially expressed MYB (myeloblastosis) and three zinc finger family TFs and signal transduction components such as calmodulin-/calcium-dependent protein kinases were observed in the Y58S comparison group. LOC_Os01g62410 (OsMYB3R-2), identified as an upregulated gene in Y58S, encodes for an MYB domain protein activation TF that regulates the CBF pathway and cell cycle progression during cold stress, resulting in increased cold tolerance (Ma et al. 2009). In a similar study, Su et al. (2010) reported the role of MYBS3 (LOC_Os10g41200) in regulating signaling pathways at low temperature, suggesting that MYB family members are good candidates for improving LTS tolerance in rice. Furthermore, molecular evidence indicates that CBF responds early, and MYBS late, to chilling stress, suggesting distinct pathways that function sequentially and complementarily to promote short- and long-term cold-stress adaptation in rice. Gene profiling on chilling-tolerant japonica rice incubated for 24 h at 10 °C revealed that an “early response” regulatory network including ROS-bZIP1 plays a crucial role in short-term adaptive responses (Yang et al. 2012). Moreover, several regulatory clusters, including bZIP factors acting on as1/ocs/TGA-like element-enriched clusters , R2R3-MYB factors acting on MYB2-like element-enriched clusters, and ERF factors acting on GCC-box/JAre-like element-enriched clusters, are involved in early chilling response, and oxidative signaling by H2O2 is at the center of the regulatory network (Yun et al. 2010).
Furthermore, genes involved in gibberellic acid (GA), indole-3-acetic acid (IAA), and cytokinin biosynthesis responded to cold temperature in such a way that their expression profiles were either downregulated or upregulated in cold-susceptible and cold-tolerant rice varieties (Park et al. 2010). For example, the IAA biosynthesis genes YUCCA1 and TAA 1:1 showed variety-specific regulation. Among the genes involved in cytokinin biosynthesis and signaling, the expression of LOG, HK1, and HK3 was significantly downregulated only in the cold-susceptible variety. Similarly, among the genes involved in ABA biosynthesis, neoxanthin synthase (NSY), and ABA-aldehyde oxidase 3 (AAO3) were downregulated only in the cold-tolerant variety. It is presumed that the levels of these bioactive hormones are maintained relatively high at cold temperatures in cold-tolerant varieties, which can help minimize the cold stress imposed on developing reproductive organs.
In a comparative transcriptome analysis of the shoots and roots of cold-tolerant variety TNG67 and cold-sensitive variety TCN1, the expression of OsRR4 type-A response regulators in roots of TNG67 was upregulated. The TFs OsIAA23, SNAC2, OsWRKY1v2, 24, 53, 71, HMGB, OsbHLH, and OsMyb were expressed in the roots or shoots of TNG67, and AP2/ERF in the shoots and roots of both varieties during cold stress, making them good candidate genes for cold-stress tolerance in rice. Also, phytohormone-related genes for ABA, polyamine, auxin, and jasmonic acid were preferentially upregulated in the shoots and roots of the cold-tolerant genotype. Functional clustering of the majority of DEGs involved in early chilling response showed their role in a complicated chilling-responsive regulatory network such as phytohormone signaling, photosynthesis pathway, ribosome translation machinery, and phenylpropanoid biosynthesis. The localization of the majority of DEGs in chloroplasts suggests a link between chilling-stress tolerance in rice and photosynthesis (Wang et al. 2016). This was observed in a comparative transcriptome profiling of the common wild rice GXWR (China)-derived chilling-tolerant chromosome segment substitution line.
4.4 Proteomics Related to LTS Tolerance
Proteins are the key components in the majority of cellular events; hence, investigating their structure, function, abundance, and interactions at a given time is advantageous to “omics” studies. Protein translational and posttranslational regulations, particularly of stressor-specific protein classes altered due to stress conditions, can also be detected by proteomics, thereby rendering a complex phenomenon such as the tolerance mechanisms for cold and other stresses well understood and addressed. Two-dimensional gel electrophoresis (2-DE) or liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) is used in protein extraction followed by protein separation and identification (Wittmann-Liebold et al. 2006; Fournier et al. 2007; Hashimoto and Komatsu 2007; Wang et al. 2013c).
Proteomic studies were undertaken pertaining to cold tolerance in rice seedlings and anthers (Cui et al. 2005; Yan et al. 2006; Hashimoto and Komatsu 2007; Zhang et al. 2014a; Lee et al. 2015). Comprehensive transcriptomic and proteomic analyses in rice have illustrated that many genes and functional proteins are involved in the crop’s chilling response (Hashimoto and Komatsu 2007; Nakashima et al. 2007; Kanneganti and Gupta 2008; Oh et al. 2009; Lee et al. 2015). Furthermore, many proteins, including otsA and otsB (trehalose synthesis), choline monooxygenase (glycine betaine synthesis), and WFT1 and WFT2 (fructan synthesis) (Garg et al. 2002; Shirasawa et al. 2006), were found to be involved in the regulation of low-temperature tolerance in rice. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) , Cui et al. (2005) observed 60 protein spots progressively upregulated in response to LTS. These cold-responsive proteins include four factors of protein biosynthesis, four molecular chaperones, two proteases, eight enzymes involved in the biosynthesis of cell wall components, and seven antioxidative enzymes and proteins linked to energy pathways as in signal transduction, besides two proteins of unknown function. In addition to these proteins, chloroplast proteome is also subject to cold stress because of the localization of identified proteins in the chloroplast. Other cold-responsive proteins are sucrose synthase (Maraña et al. 1990), phenylalanine ammonia-lyase (Sanchez-Ballesta et al. 2000), and ferritin (Kawamura and Uemura 2003). Proteins involved in energy metabolism in the leaf blade were upregulated, while defense-related proteins were downregulated or even disappeared when rice seedlings were exposed to 5 °C for 48 h (Hashimoto and Komatsu 2007). Hashimoto and Komatsu (2007) used the 2D PAGE-based proteomics method for rice root plasma membrane and identified 12 cold-responsive proteins, including cold shock protein-1, which decreased significantly under cold-stress conditions. Most of the cold-responsive proteins associated with energy production, signal transduction, protein synthesis, and defense were revealed by their functional characterization.
Proteins involved in phytohormone biosynthesis play a role in stress tolerance. Asakura et al. (2004) found that GA-related proteins have increased expression during rice seed germination. The elevated expression of GA receptor GID1L2 observed in the resistant strain suggests the role played by GA in mediating the response to cold temperature in the germinating embryo. The low-temperature germination of the resistant rice line was associated with proteins involved in GA signaling, protein trafficking, and ABA-mediated stress response compared with the susceptible strain. Colebrook et al. (2014) also noticed that increased GA biosynthesis and GA signaling were linked to stress tolerance. The elevated expression of TF HBP-1b involved in the general mechanism of protein trafficking through a secretory pathway and two proteins of unknown function, UPF0041 domain-containing protein (LOC_Os07g26700.1) and the expressed protein (LOC_Osg09910.1), suggested their possible role in seed germination or cold stress (Lee et al. 2015). Phosphorylation of cellular proteins or protein kinase activation during the initial stage of cold acclimation has also been observed (Garbarino et al. 1991), and a fragment of RuBisCO large subunit protein was phosphorylated to a greater extent than others in cold-tolerant rice varieties using 2-DE analysis (Komatsu et al. 1999).
Gel-based protein separation has been used in proteomic studies, which has resulted in the identification of only high-abundant proteins, leaving TFs, kinases, and transport proteins belonging to low-abundant protein classes undetected. Although advanced LC-based separation techniques have resulted in the detectability of low-abundant proteins, phosphorylation, glycosylation, and oxidation caused by posttranslational changes that are likely to be induced by stressors are yet to be comprehensively explained with the use of these approaches. Also, metabolic processes mediated by plant hormones as well as subcellular protein translocation and protein-protein interactions are yet to be found associated with the cold-stress phenomenon and tolerance mechanisms. To move forward with our knowledge on a time-dependent response, new proteomic strategies, such as hydrogen-deuterium exchange and surface plasmon resonance-MS, together with integrated cell biology approaches such as immune precipitation and live imaging analysis, will be required.
4.5 Metabolomics Related to LTS Tolerance
Metabolites are the final response of a biological system to environmental changes so that an aberrant metabolism is linked to the most predictive of phenotypes. The successful application of metabolomics can provide a deeper insight into a plant’s phenotypic response to abiotic stresses and can determine the pattern related to stress tolerance. For example, in a targeted metabolite analysis of two rice genotypes under LTS (13/12 °C), Morsy et al. (2007) observed that the chilling-tolerant genotype exhibited accumulated galactose and raffinose, while the same sugars were found to be decreasing in the chilling-sensitive variety. Also, a higher endogenous content of oxidative products and the presence of a more efficient reactive oxygen species (ROS)-scavenging metabolism were found in the chilling-tolerant genotype during chilling stress. Similarly, Arbona et al. (2013) studied photosynthetic dysfunction and effectors of osmotic readjustment at primary metabolite levels (sugars, amino acids, and Krebs cycle intermediates) and secondary levels (antioxidants) and observed a relative accumulation of some primary or secondary metabolites.
Most sugars have earlier been reported to function as osmoprotectants, nutrients, and signaling molecules in rice (Guy et al. 2008; Ma et al. 2009). Similar studies have been carried out in other crops (Urano et al. 2009; Bowne et al. 2012; Araújo et al. 2013). In a comparative metabolomics study between the varieties LTH (cold-tolerant japonica) and IR29 (cold-sensitive indica) under no-stress and chilling-stress conditions (4 °C for 2, 8, 24, and 48 h, and recovery of 24 h), it was observed that 82 of 106 metabolites exhibited significant differences and described 18.1% of the total PV (Zhao et al. 2013). Of the total of 120 stress vs. control comparisons, 85 (71%) cases had significantly increased amino acid levels, whereas only 7 (6%) cases had significantly decreased amino acid levels, which involved aspartic acid, cysteine, glutamic acid, and glycine. Compared to IR29, LTH recorded more amino acids that significantly increased at all times of the stress as well as considerably higher levels of cysteine, isoleucine, phenylalanine, proline, serine, threonine, and valine. This strongly suggests that differential amino acid accumulation is a general feature of the variety in response to chilling stress at the seedling stage. Regarding organic acids, 81 of 248 (33%) showed significantly decreased levels of LTH compared to IR29, whereas only 24 (10%) cases had significantly increased levels. Consistent decreased levels were obtained for four organic acids (oleic, quinic, eicosanoic, and sinapic) across all times of the stress in both genotypes, implying that energy production is remarkably inhibited in rice during chilling stress. Furthermore, 34 of 304 (11%) cases showed significantly increased levels of some sugars in LTH at later times of the stress, signifying that these late-accumulating sugars may be associated with the cold tolerance of LTH. Zhang et al. (2016) performed a comparative metabolomics study between japonica Nipponbare and indica 93-11 at six times during chilling treatment and found that amino acid accumulation occurred on a large scale and was consistent with the appearance of chilling injury. The accumulation of antioxidation-related compounds appeared earlier in Nipponbare than in 93-11 at the mid-treatment stage, whereas, during recovery, a higher level of ROS was observed in Nipponbare. Furthermore, metabolites related to stress tolerance and senescence were found to be induced/accumulated in Nipponbare and 93-11, respectively.
The combinational approaches of genome and metabolome are more interesting for phenotype prediction and are quite useful in breeding for stress tolerance. Selection based solely on genetic markers is highly biased because the environment profoundly influences most of the economic traits. The development of a metabolite quantitative trait locus (mQTL) and metabolome-wide association studies (MWAS) could be helpful in crop improvement by overcoming the problems emerging from differing environmental conditions during selection. This field will take advantage of the new plant genomes issued recently and of the modern and more powerful metabolite profiling tools (Dumas 2012; Wei et al. 2018). Some useful results at the metabolome level and the involvement of different metabolites in plant responses, as discussed above, have provided more insight into the complexity of cold-stress response. Even though this has extended our understanding of the molecular mechanism of plant response to stresses, their reconfiguration under stresses is still quite complex because of the involvement of multiple molecular pathways (Guy et al. 2008).
5 Breeding Approaches for LTS Tolerance in Rice
Developing breeding strategies for LTS-tolerant rice varieties is still a challenge to plant breeders because of the complex nature and lack of suitable rice varieties for high-latitude regions, mainly in China, Japan, Australia, and Korea. Therefore, identifying LTS-tolerance traits and breeding tolerant rice varieties are necessary for these regions. Several researchers have proposed different strategies such as adjusting sowing time, selecting suitable rice varieties with growth duration that avoids the peak LTS periods, and replacing LTS-susceptible rice varieties (Ye et al. 2009). However, with rapid changes in GCVs and the expected population increases, breeding for LTS tolerance and improving LTS-tolerance mechanisms are the critical factors to meet future global food demand.
5.1 Improving LTS Tolerance by Conventional Breeding Approaches
Cold tolerance is the ability of rice plants to sustain yield in the presence of low-temperature stress (Shakiba et al. 2017). Genetic breeding has been used as the approach to cope with low-temperature sensitivity. The indica subspecies are better adapted to tropical environments such as India, China, and Indonesia, while japonica cultivars have more adaptation under temperate climates such as those in Japan, Korea, and Java, Indonesia (Takahashi 1984). It has been observed that japonica genotypes are relatively better in tolerating a higher degree of cold stress at the germination stage (Lee 2001; da Cruz and Milach 2004) as well as at the vegetative and reproductive stages (Saito et al. 2004; da Cruz et al. 2006a; Zhang et al. 2017; Xiao et al. 2018). However, some indica genotypes from high-latitude regions may have moderate cold tolerance (Gautam et al. 2018). There are reports of some javanica, an ecotype of japonica, being tolerant of cold (Sweeney and McCouch 2007). Cold-tolerance genes from javanica cultivars such as Silewah, Lambayeque 1, and Padi Labou Alumbis were introduced into several temperate japonica breeding lines in Japan (Saito et al. 2004).
Selection under field conditions for cold tolerance in rice is unpredictable; hence, for effective selection, robust screening protocols using strong selective agents such as low temperature and the use of controlled air or low water temperature are highly important (da Cruz et al. 2006b). The major limitation to evaluating large plant populations in controlled-temperature environments is to provide enough space for the material. Growth chambers and phytotron facilities lead to quicker and more precise results; however, small-sample testing leads to a loss in effective population size. To deal with these limitations, some rice breeding programs use cold water under field conditions as a selection criterion, allowing the evaluation of many different populations and thousands of plants per population (Snell et al. 2008). Several experimental stations in Japan (Okamoto et al. 1986; Horisue et al. 1988; Nagano 1998; Shinada et al. 2013), Bangladesh (Khatun et al. 2016), and Korea (Jeong et al. 1998) have successfully used cold water to screen rice breeding material for cold tolerance. Different traits depend on the developmental stage, which is used as an indicator to identify cold-tolerant/susceptible lines. Some correlation studies revealed that varieties having germination and seedling tolerance under low-temperature conditions might also be tolerant at the booting and flowering stages (Ye et al. 2009). Inheritance and heritability of LTS tolerance at the germination stage showed involvement of both additive and nonadditive gene actions, with the latter component relatively more important for coleoptile length and coleoptile growth decrease. In a similar kind of study, epistatic interaction (a nonadditive effect) was found to be important for rice germination capacity at low temperature. Some tolerance genes may be more important, whereas others are stage-specific. No such genes have been demonstrated, which are responsible for cold tolerance over all the stages.
Selection for cold-tolerant genotypes in rice has shown a greater success rate due to high heritability estimates for low-temperature germinability (Sthapit and Witcombe 1998; da Cruz et al. 2006b). The nature and magnitude of gene action involved in different traits such as root and shoot length and plant stature are used as indicators under cold stress, which helps in devising a breeding program for developing cold-tolerant genotypes (Datta and Siddiq 1983; Kaw and Khush 1986; Acharya 1987). Involvement of two major genes, Cts1 and Cts2, has been reported at the vegetative stage responsible for cold tolerance and estimated through leaf yellowing (Kwak 1984) and withering (Nagamine 1991). On the other hand, studies of Andaya and Tai (2006) reported cold tolerance at the vegetative stage as a complex trait involving multiple genes and, similarly, several major genes are involved in cold tolerance at the reproductive stage. Studies on correlating cold tolerance with morphological traits in temperate japonica varieties show that tolerance is governed by four or more loci, which were shown to be linked to morphological marker genes (Futuhara and Toriyama 1966). It has been reported that several cold-tolerant cultivars have already been released in different countries despite the complex genetic basis and difficulties and limitations of selection for cold tolerance (da Cruz et al. 2013). These varieties may serve as a useful genetic repository rather than gene donors to develop cold tolerance in new rice varieties, which are lacking the trait. Much work on trait improvement resulted in 57% of the rice varieties in Korea having tolerance, and they are highly tolerant of low temperatures (Lee 2001). Each country developed its own strategy for breeding for cold tolerance. However, the main advances have been obtained within japonica cultivars. Therefore, the challenge remains to develop indica-type cultivars with adequate cold tolerance for high-latitude regions (Bierlen et al. 1997). An apparently simple solution could be to cross indica genotypes with japonica ones to transfer genes for cold tolerance from japonica. However, the differences between these two rice groups make it difficult to maintain desirable indica characteristics, such as the cooking quality needed for consumer acceptance. Also, gene introgression from indica to japonica rice has shown problems of high sterility and poor plant type with some linkage drag in the progenies (Khush 2005). Furthermore, overcoming the major constraints associated with conventional breeding for cold tolerance in rice has been of limited success with gene transfer from wild relatives (Flowers and Yeo 1995). Another difficulty in the selection of cold-tolerant varieties in field conditions is the lack of appropriate selection pressure, which provides critical stress-environment control (Blum 1988). Conventional breeding is an extremely slow process for generating varieties with improved tolerance of stress conditions. In addition, incompatibility in wider crosses along with limited germplasm resources for stress tolerance is a major limitation encountered in conventional breeding. Therefore, integrating traditional breeding programs with modern compatible molecular methods and elucidating the genetic mechanisms of this complex phenomenon will improve precision and speed in the development of genotypes with low-temperature tolerance.
5.2 Improving LTS Tolerance by Selective Introgression
The selective introgression breeding (SIB) strategy is a powerful tool for the simultaneous improvement of tolerance and also to dissect complex traits. This is based on two approaches , such as developing a larger number of trait-specific introgression lines via early backcross breeding and using a marker-facilitated approach to track the gene flow from donors to recipients. In addition to that, the selected early backcross breeding population can be used for identifying the genomic regions for the association of target traits of interest through QTL mapping (Pang et al. 2017; Feng et al. 2018; Jewel et al. 2018). In the Green Super Rice (GSR) breeding program, this SIB strategy has laid the foundation for a better molecular and genetic understanding of complex traits in rice. This methodology provides more advantages than classical QTL mapping methods, such as decreasing the cost in genotyping and phenotyping with selected lines, high statistical power to detect QTLs for targeted traits, and the selected lines are expected to carry the beneficial alleles of QTLs (Ali et al. 2017; Liang et al. 2018). By dissecting complex traits and developing trait-specific introgression lines, Liang et al. (2018) successfully identified cold-tolerance QTLs on chromosomes 1, 2, 3, 4, 6, 9, 11, and 12. A total of 17 QTLs for cold tolerance at the reproductive stage were detected in 84 cold-tolerant introgression lines (ILs) selected from five BC2F4 populations in a Chaoyou genetic background using a consensus linkage map. In addition, 310 random ILs from the same BC populations were used for dissecting the genetic networks underlying cold tolerance by detecting QTLs and functional genetic units (FGUs). This study led to the discovery of QTLs qCT3.12, qCT6.7, and qCT9.6 that were validated in random BC populations. A QTL for LTG was fine-mapped by Shim et al. (2019) with the help of two introgression lines, TR5 and TR20, which were crossed to common parent Hwaseong to develop F2:3 populations. qLTG1 was located in a 167-kb region between two SSR markers (RM10310 and RM10326) and was found to harbor 18 genes , with nine of them annotated with specific gene functions. The allelic effect at the qLTG1 locus was contributed by Oryza rufipogon that was observed to increase LTG and spikelets per panicle.
5.3 Improving LTS Tolerance by Genetic Transformation
Transgenic approaches can be used to improve cold tolerance in rice. These methods are promising, wherein improvement can be made through the introduction from across trans-species or through the disruption of specific DNA sequences using RNAi technology. Gene expression analysis has made clear the physiological mechanisms responsible for tolerance against various abiotic stresses, including low temperature, in plants (Gao et al. 2008; Pan et al. 2020; Xu et al. 2020). Several chilling stress-responsive genes after their isolation and characterization have been found to encode proteins that act as enzymes for the biosynthesis of osmoprotectants. Transcription factors, especially those from the CBF/DREB1 family, play a more important role (Wang et al. 2008; Zhang et al. 2009; Su et al. 2010). Using the integrated approach of T-DNA-tagged rice plants and inverse PCR, many genes involved with several metabolic pathways were detected and characterized at the molecular level. When plants are exposed to cold stress, they show changes at the gene expression level, and the products of cold-inducible genes may either directly protect against cold stress or further regulate the expression of other genes (Yamaguchi-Shinozaki and Shinozaki 2004; Chen et al. 2008; Su et al. 2010). Several transgenic rice lines or overexpressed lines (OX) have been developed, which are responsible for the overexpression of cold-inducible genes, usually encoding TFs for the transcriptional regulation of these genes. The upregulated or overexpressed important genes/TFs with respect to their function, cold stress condition, the phenotype of OX lines, and side effects in OX lines, if any, in comparison to nontransformed (WT) plants, are described in Table 5. Overexpression of OsCTZFP8 in transgenic rice exhibited more cold-tolerant phenotypes than nontransgenic control plants by showing higher pollen fertility and seed-setting rate (Jin et al. 2018). The significant overexpression of cold-responsive genes and TFs suggests a practical utility of these genes at the field level requiring a systematic assessment, and this needs further use in crop improvement.
5.4 Improving LTS Tolerance by Genome Editing
Emerging advanced genome editing tools (GETs) such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), meganucleases (MNs), and CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein) play a greater role in the understanding of the various biological functions by manipulating the desired target gene of interest and also providing new opportunities to create genetic diversity in various crops (Vats et al. 2019; Wolter et al. 2019). However, these ZFNs, TALENs, and MNs are so expensive in the cloning procedure, have low target specificity, and are time-consuming and laborious, whereas the CRISPR/Cas system is a cheap, easy-to-design, and unique tool for precise and efficient genome editing at the single-base level. CRISPR/Cas9 involves mainly single-guided RNA (sgRNA) in contrast to ZFNs and TALENs and provides mutagenesis at high frequency, thus increasing the number of recombination sites and target specificity in plant genomes (Jaganathan et al. 2018; Molla and Yang 2019). Currently, using this technology, several researchers have targeted genome editing for biotic and abiotic stress tolerance and also grain quality-related genes. Over the past decade, CRISPR/Cas9 has been widely used to produce novel rice varieties with improved target traits such as grain weight, panicle architecture, aroma, high amylose content, and tolerance of bacterial blight and blast, drought, and cold (Mishra et al. 2018; Fiaz et al. 2019; Jun et al. 2019). For instance, Li et al. (2016) edited four grain yield-related genes (Gn1a, DEP1, GS3, and IPA1) and Xu et al. (2016) edited three genes related to grain weight (GW2, GW5, and TGW6) using the CRISPR/Cas9 system. They noticed a significant increase in phenotypic traits, such as increasing grain number and size, and dense erect panicles. In response to cold stress in rice, the CRISPR/Cas9 system was used to edit two TFs, TIFY1a (LOC_Os03g47970) and TIFY1b (LOC_Os03g52450), in rice, revealing single-base-pair insertion and deletion and also long fragment deletion. Thus, employing CRISPR/Cas9 technology to investigate the role of TIFY1a and TIFY1b might reveal a novel pathway that controls cold adaptation in rice (Huang et al. 2017). CBFs have been studied in response to cold tolerance through genome editing, which has shown that CBF triple mutants were extremely sensitive to cold acclimation-dependent freezing stress (Jia et al. 2016).
Breeding for LTS using conventional methods has met with limited success in improving tolerant rice varieties because of the lack of efficient selection criteria, the time-consuming work, and required expensive facilities for screening, and it also involves a multigenic trait. Therefore, an alternative breeding strategy such as genomic-assisted breeding and gene transformation technologies can provide a viable option to improve LTS tolerance in rice. The ability of the CRISPR/Cas9 system to generate transgene-free genome-modified plants, create site-specific and SNP mutations, and make large-scale changes in chromosome structure at the single and multicellular levels provides a comprehensive target trait mechanism and more helpful breeding strategy to enhance biotic and abiotic stress tolerance in rice. The successful editing of important genes related to improving grain yield and quality traits has proven that this CRISPR/Cas9 technology could be a promising tool for understanding the molecular and physiological functional aspects of genes and TFs that influence target traits.
6 Conclusions and Future Prospects
This chapter has mainly focused on LTS-tolerance traits and associated genetics in rice, which have significant importance in tropical and temperate regions across the globe for increasing yield productivity and sustainability. The LTS tolerance-related QTLs, genes, enzymes, and their interactions are involved in stress-tolerance mechanisms in different growth stages. In order to understand the interaction of genetics, physiological and metabolic responses to plants need to be analyzed using a growth-stage specific and precise phenotypic characterization. The LTS-tolerance trait associated with QTLs and genes will be useful in designing molecular breeding strategies for improving LTS tolerance in rice varieties. The combinational approaches of genomics and metabolomics are more interesting for phenotype prediction and are quite useful in breeding for stress tolerance. Selection based solely on genetic markers is highly biased because the environment profoundly influences most of the economic traits. The development of a metabolomics quantitative trait locus and metabolome-wide association studies could be helpful in crop improvement by overcoming the problems emerging from differing environmental conditions during selection. This field will take advantage of the new plant genomes issued recently and of the modern and more powerful metabolite profiling tools (Dumas 2012; Wei et al. 2018). Some useful results at the metabolome level and the involvement of different metabolites in plant responses, as discussed above, have provided more insight into the complexity of cold-stress response. Even though this has extended our understanding of the molecular mechanisms of plant response to stresses, their reconfiguration under stresses is still quite complex because of the involvement of multiple molecular pathways (Guy et al. 2008).
However, in a breeding program for the development of LTS tolerance, advances in omics-related technologies have led to the development of well-planned phenotypic experiments that offer deeper insight into gene function along with gene effects on the phenotype in a specified biological context. There is a significant positive correlation of LTS-tolerance traits among the different growth stages of rice, and this suggests that rice varieties with high germination rate and early seedling vigor-related traits under LTS conditions are also likely to be more tolerant in the reproductive stage (Ye et al. 2009). It is reasonable to consider that LTS-tolerant genotypes in the GS, SS, and RS may rely on the diverse genetic components of QTLs and genes on different chromosomes that are ensuring LTS tolerance. The rapid development of advanced genome sequencing and larger sets of polymorphic SNPs help in identifying potential candidate genes. Based on the comprehensive literature survey under LTS regarding GS-, SS-, and RS-specific traits and associated with fine-tuned and colocalized stage-specific QTLs (in GS and SS), the highest numbers of QTLs were located on chromosomes 1 and 7 (Pan et al. 2015; Shakiba et al. 2017; Zhang et al. 2018; Sun et al. 2019).
Similarly, in three combinations of stage-specific QTLs (GS, SS, and RS) associated with multiple traits on chromosome 10 (Li et al. 2013; Pan et al. 2015; Schläppi et al. 2017), seven genetic regions are colocalized with candidate genes and QTLs on four chromosomes (1, 3, 5, and 6), and this provides insights into identifying the candidate genes and developing functional allele-specific markers for improving LTS tolerance in breeding programs. The concept of omics studies has amassed a great deal of information at the transcript, protein, and metabolite levels to perceive the tolerance mechanisms of plants under stress. To fully understand the complex regulatory nature of plants against stresses, a highly coordinated approach such as systems biology needs to be identified. Unfortunately, the integration of data outputs from phenomes, transcriptomes, proteomes, and metabolomes has been found to be inefficient and ineffective yet, despite the marvelous progress shown in bioinformatics. However, future approaches need to integrate these through robust computing platforms that should be able to predict the active biochemical and molecular genetic networks to exploit LTS tolerance in rice breeding. This could potentially lead to detecting and identifying master stress regulators to target the right biomarkers for improving rice LTS tolerance effectively.
References
Acharya S (1987) Genetic parameters and their implication in breeding cold tolerant varieties of rice (Oryza sativa L.). Crop Improv 14:100–103
Ali AJ, Xu JL, Ismail AM et al (2006) Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crop Res 97:66–76
Ali J, Xu J-L, Gao Y-M et al (2017) Harnessing the hidden genetic diversity for improving multiple abiotic stress tolerance in rice (Oryza sativa L.). PLoS One 12:e0172515
Almeida DM, Almadanim MC, Lourenço T et al (2016) Screening for abiotic stress tolerance in rice: salt, cold, and drought. In: Environmental responses in plants. Springer, New York, NY, pp 155–182
Andaya V, Mackill D (2003a) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica × indica cross. Theor Appl Genet 106:1084–1090
Andaya VC, Mackill DJ (2003b) Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J Exp Bot 54:2579–2585. https://doi.org/10.1093/jxb/erg243
Andaya VC, Tai TH (2006) Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor Appl Genet 113:467–475
Araújo WL, Trofimova L, Mkrtchyan G et al (2013) On the role of the mitochondrial 2-oxoglutarate dehydrogenase complex in amino acid metabolism. Amino Acids 44:683–700
Arbona V, Manzi M, de Ollas C, Gómez-Cadenas A (2013) Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int J Mol Sci 14:4885–4911
Asakura T, Nakaizumi T, Hirose S et al (2004) Proteome analysis of the regulation of rice seed germination. In: Plant and cell physiology. Oxford University Press, Oxford, pp s148–s148
Bai B, Wu J, Sheng W-T et al (2015) Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress. Int J Mol Sci 16:11398–11416
Basuchaudhuri P (2014) Cold tolerance in rice cultivation. CRC Press, Boca Raton, FL
Bertin P, Kinet JM, Bouharmont J (1996) Evaluation of chilling sensitivity in different rice varieties: relationship between screening procedures applied during germination and vegetative growth. Euphytica 89:201–210
Bevilacqua CB, Basu S, Pereira A et al (2015) Analysis of stress-responsive gene expression in cultivated and weedy rice differing in cold stress tolerance. PLoS One 10:e0132100
Bierlen R, Wailes EJ, Cramer GL (1997) The mercosur rice economy. Ark Exp Stat Bull 954:1–58
Biswas PS, Khatun H, Das N et al (2017) Mapping and validation of QTLs for cold tolerance at seedling stage in rice from an indica cultivar Habiganj Boro VI (Hbj.BVI). 3 Biotech 7:359. https://doi.org/10.1007/s13205-017-0993-1
Blum A (1988) Plant breeding for stress environments. CRC Press, Boca Raton, FL. 223 p
Board JE, Peterson ML, Ng E (1980) Floret sterility in rice in a cool environment. 1. Agron J 72:483–487
Bonnecarrère V, Borsani O, Díaz P et al (2011) Response to photoxidative stress induced by cold in japonica rice is genotype dependent. Plant Sci 180:726–732
Bonnecarrere V, Quero G, Monteverde E et al (2015) Candidate gene markers associated with cold tolerance in vegetative stage of rice (Oryza sativa L.). Euphytica 203:385–398
Bowne JB, Erwin TA, Juttner J et al (2012) Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol Plant 5:418–429
Chen L, Qiaojun L, Sun ZX et al (2006a) QTL mapping of low temperature germinability in rice. Zhongguo Shuidao Kexue 20:159–164
Chen L, Qiaojun L, Sun ZX et al (2006b) QTL mapping of low temperature on germination rate of rice. Rice Sci 13:93–98
Chen JQ, Meng XP, Zhang Y et al (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30:2191–2198. https://doi.org/10.1007/s10529-008-9811-5
Chen NA, Xu Y, Wang XIN et al (2011) OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ 34:52–64
Chen J, Lin T, Xu H et al (2012) Cold-induced changes of protein and phosphoprotein expression patterns from rice roots as revealed by multiplex proteomic analysis. Plant Omics 5:194
Cheng L, Wang J, Uzokwe V et al (2012) Genetic analysis of cold tolerance at seedling stage and heat tolerance at anthesis in rice (Oryza sativa L.). J Integr Agric 11:359–367
Chinnusamy V, Ohta M, Kanrar S et al (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Chinnusamy V, Zhu J, Zhu J-K (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451
Chinnusamy V, Zhu JK, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. In: Plant stress tolerance. Springer, New York, NY, pp 39–55
Chou WL, Huang LF, Fang JC et al (2014) Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa. Plant Mol Biol 85:443–458. https://doi.org/10.1007/s11103-014-0196-7
Colebrook EH, Thomas SG, Phillips AL, Hedden P (2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217:67–75
da Cruz RP, Milach SCK (2000) Breeding for cold tolerance in irrigated rice. Ciência Rural 30:909–917
da Cruz RP, Milach SCK (2004) Cold tolerance at the germination stage of rice: methods of evaluation and characterization of genotypes. Sci Agric 61:1–8
da Cruz RP, Milach SCK, Federizzi LC (2006a) Rice cold tolerance at the reproductive stage in a controlled environment. Sci Agric 63:255–261
da Cruz RP, Milach SCK, Federizzi LC et al (2006b) Inheritance of rice cold tolerance at the germination stage. Genet Mol Biol 29:314–320
da Cruz RP, Duarte ITL, Cabreira C (2010) Inheritance of rice cold tolerance at the seedling stage. Sci Agric (Piracicaba, Braz) 67:669–674. https://doi.org/10.1590/S0103-90162010000600008
da Cruz RP, Sperotto RA, Cargnelutti D et al (2013) Avoiding damage and achieving cold tolerance in rice plants. Food Energ Secur 2:96–119. https://doi.org/10.1002/fes3.25
Cui S, Huang F, Wang J et al (2005) A proteomic analysis of cold stress responses in rice seedlings. Proteomics 5:3162–3172
Cui Y, Zhang F, Zhou Y (2018) The application of multi-locus GWAS for the detection of salt-tolerance loci in rice. Front Plant Sci 9:1464
Dai L, Lin X, Ye C et al (2004) Identification of quantitative trait loci controlling cold tolerance at the reproductive stage in Yunnan landrace of rice, Kunmingxiaobaigu. Breed Sci 54:253–258
Datta D, Siddiq EA (1983) Genetic analysis of cold tolerance at seedling phase in rice. Indian J Genet Plant Breed 43:345–349
Ding Y, Shi Y, Yang S (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol 222:1690–1704
Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21:972–984
Donoso G, Cabas P, Paredes M et al (2015) Cold tolerance evaluation of temperate rice (Oryza sativa L. ssp. japonica) genotypes at seedling stage. Gayana Bot 72:1–13
Dumas M-E (2012) Metabolome 2.0: quantitative genetics and network biology of metabolic phenotypes. Mol BioSyst 8:2494–2502
Farrell T, Fox KM, Williams RL et al (2001) Temperature constraints to rice production in Australia and Laos: a shared problem. In: Increased lowland rice production in the mekong region. Australian Centre for International Agricultural Research, Canberra, ACT, pp 129–137
Feng B, Chen K, Cui Y et al (2018) Genetic dissection and simultaneous improvement of drought and low nitrogen tolerances by designed QTL pyramiding in rice. Front Plant Sci 9:306. https://doi.org/10.3389/fpls.2018.00306
Fiaz S, Ahmad S, Noor MA et al (2019) Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities. Int J Mol Sci 20:888
Flowers TJ, Yeo AR (1995) Breeding for salinity resistance in crop plants: where next? Funct Plant Biol 22:875–884
Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP (2007) Multidimensional separations-based shotgun proteomics. Chem Rev 107:3654–3686
Fujino K, Matsuda Y (2010) Genome-wide analysis of genes targeted by qLTG3-1 controlling low-temperature germinability in rice. Plant Mol Biol 72:137
Fujino K, Sekiguchi H (2011) Origins of functional nucleotide polymorphisms in a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Plant Mol Biol 75:1–10
Fujino K, Sekiguchi H, Sato T et al (2004) Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor Appl Genet 108:794–799
Fujino K, Sekiguchi H, Matsuda Y et al (2008) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci U S A 105:12623–12628. https://doi.org/10.1073/pnas.0805303105
Fursova OV, Pogorelko GV, Tarasov VA (2009) Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429:98–103
Futuhara Y, Toriyama K (1966) Genetic studies on cool tolerance in rice: III. Linkage relations between genes controlling cool tolerance and marker genes of NAGAO and TAKAHASHI. Jpn J Breed 16:231–242
Gao JP, Chao DY, Lin HX (2008) Toward understanding molecular mechanisms of abiotic stress responses in rice. Rice 1:36–51
Garbarino JE, Hurkman WJ, Tanaka CK, DuPont FM (1991) In vitro and in vivo phosphorylation of polypeptides in plasma membrane and tonoplast-enriched fractions from barley roots. Plant Physiol 95:1219–1228
Garg AK, Kim J-K, Owens TG et al (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci U S A 99:15898–15903
Gautam A, Suresh J, Gudade BA et al (2018) Rice: grappling with cold under climatic changes, global impact and counter strategies. Adv Res 13:1–9
Ge L-F, Chao D-Y, Shi M et al (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228:191–201
Ghosh T, Rai M, Tyagi W, Challam C (2016) Seedling stage low temperature response in tolerant and susceptible rice genotypes suggests role of relative water content and members of OsSNAC gene family. Plant Signal Behav 11:1–5. https://doi.org/10.1080/15592324.2016.1138192
Gilmour SJ, Sebolt AM, Salazar MP et al (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124:1854–1865
Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54:767–781
Gothandam KM, Nalini E, Karthikeyan S, Shin JS (2010) OsPRP3, a flower specific proline-rich protein of rice, determines extracellular matrix structure of floral organs and its overexpression confers cold-tolerance. Plant Mol Biol 72:125
Greco M, Chiappetta A, Bruno L, Bitonti MB (2012) In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J Exp Bot 63(2):695–709
Gupta BB, Salgotra RK, Bali AS (2009) Status paper on rice in Jammu and Kashmir. Rice Knowledge Management Portal (RKMP), Hyderabad. http://www.rkmp.co.in/
Gururani MA, Venkatesh J, Ganesan M et al (2015) In vivo assessment of cold tolerance through chlorophyll-a fluorescence in transgenic zoysiagrass expressing mutant phytochrome A. PLoS One 10:1–17. https://doi.org/10.1371/journal.pone.0127200
Guy C, Kaplan F, Kopka J et al (2008) Metabolomics of temperature stress. Physiol Plant 132:220–235
Han LZ, Qiao YL, Cao GL et al (2004) QTLs analysis of cold tolerance during early growth period for rice. Rice Sci 11:245–250
Han LZ, Zhang YY, Qiao YL et al (2006) Genetic and QTL analysis for low-temperature vigor of germination in rice. Acta Genet Sin 33:998–1006
Hashimoto M, Komatsu S (2007) Proteomic analysis of rice seedlings during cold stress. Proteomics 7:1293–1302
Horisue N, Kunihiro Y, Higashi T et al (1988) Screening for cold tolerance of Chinese and Japanese rice varieties and selection of standard varieties. Trop Agric Res Ser 21:76–87
Hou MY, Jiang L, Wang CM, Wan JM (2003) Detection and analysis of QTLs for low temperature germinability in rice (Oryza sativa L.). Rice Genet Newsl 20:52–55
Hu H, You J, Fang Y et al (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Huang J, Sun S, Xu D et al (2012) A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol Biol 80:337–350
Huang X, Zeng X, Li J, Zhao D (2017) Construction and analysis of tify1a and tify1b mutants in rice (Oryza sativa) based on CRISPR/Cas9 technology. J Agric Biotechnol 25:1003–1012
Hussain HA, Hussain S, Khaliq A et al (2018) Chilling and drought stresses in crop plants: implications, cross talk, and potential management opportunities. Front Plant Sci 9:393
Ito Y, Katsura K, Maruyama K et al (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153. https://doi.org/10.1093/pcp/pci230
Jaganathan D, Ramasamy K, Sellamuthu G et al (2018) CRISPR for crop improvement: an update review. Front Plant Sci 9:985
Jena KK, Kim SM, Suh JP et al (2012) Identification of cold-tolerant breeding lines by quantitative trait loci associated with cold tolerance in rice. Crop Sci 52:517–523
Jeong EG, Baek MK, Yea JD et al (1998) Screening of characters related to cold tolerance of Korean leading rice varieties. RDA J Crop Sci 40:25
Jewel Z, Ali J, Pang Y et al (2018) Developing green super rice varieties with high nutrient use efficiency by phenotypic selection under varied nutrient conditions. Crop J 7:368–377. https://doi.org/10.1016/j.cj.2019.01.002
Ji SL, Jiang L, Wang YH et al (2008) QTL and epistasis for low temperature germinability in rice. Acta Agron Sin 34:551–556
Ji H, Kim SR, Kim YH et al (2010a) Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J 61:96–106. https://doi.org/10.1111/j.1365-313X.2009.04039.x
Ji ZJ, Zeng YX, Zeng DI et al (2010b) Identification of QTLs for rice cold tolerance at plumule and 3-leaf-seedling stages by using QTLNetwork software. Rice Sci 17:282–287. https://doi.org/10.1016/S1672-6308(09)60028-7
Ji X, Dong B, Shiran B et al (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol 156:647–662
Jia Y, Ding Y, Shi Y et al (2016) The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol 212:345–353
Jiang L, Xun M, Wang J, Wan J (2008) QTL analysis of cold tolerance at seedling stage in rice (Oryza sativa L.) using recombination inbred lines. J Cereal Sci 48:173–179
Jiang Y, Cai Z, Xie W et al (2012) Rice functional genomics research: progress and implications for crop genetic improvement. Biotechnol Adv 30:1059–1070. https://doi.org/10.1016/j.biotechadv.2011.08.013
Jiang N, Shi S, Shi H et al (2017) Mapping QTL for seed germinability under low temperature using a new high-density genetic map of rice. Front Plant Sci 8:1–9. https://doi.org/10.3389/fpls.2017.01223
Jin YM, Piao R, Yan YF et al (2018) Overexpression of a new zinc finger protein transcription factor OsCTZFP8 improves cold tolerance in rice. Int J Genom 23:5480617
Joo J, Lee YH, Kim Y et al (2013) Abiotic stress responsive rice ASR1 and ASR3 exhibit different tissue-dependent sugar and hormone-sensitivities. Mol Cell 35:421–435. https://doi.org/10.1007/s10059-013-0036-7
Jun REN, Xixun HU, Kejian W, Chun W (2019) Development and application of CRISPR/Cas system in rice. Rice Sci 26:69–76
Kanneganti V, Gupta AK (2008) Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol Biol 66:445–462
Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45:346–350
Kaw RN, Khush GS (1986) Combining ability for low-temperature tolerance in rice. In: Rice genetics I (in two parts). World Scientific, Singapore, pp 593–612
Kawamura Y, Uemura M (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36:141–154
Khatun H, Biswas PS, Hwang HG, Kim K-M (2016) A quick and simple in-house screening protocol for cold tolerance at seedling stage in rice. Plant Breed Biotechnol 4:373–378
Khush GS (2005) What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol 59:1–6
Kim S-I, Tai TH (2011) Evaluation of seedling cold tolerance in rice cultivars: a comparison of visual ratings and quantitative indicators of physiological changes. Euphytica 178:437–447
Kim S-I, Tai TH (2013) Identification of SNPs in closely related Temperate Japonica rice cultivars using restriction enzyme-phased sequencing. PLoS One 8:e60176
Kim S-H, Kim J-Y, Kim S-J et al (2007) Isolation of cold stress-responsive genes in the reproductive organs, and characterization of the OsLti6b gene from rice (Oryza sativa L.). Plant Cell Rep 26:1097–1110
Kim S-J, Lee S-C, Hong SK et al (2009) Ectopic expression of a cold-responsive OsAsr1 cDNA gives enhanced cold tolerance in transgenic rice plants. Mol Cell 27:449–458
Kim S-MM, Suh J-PP, Lee C-KK et al (2014) QTL mapping and development of candidate gene-derived DNA markers associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Gen Genomics 289:333–343. https://doi.org/10.1007/s00438-014-0813-9
Koh S, Lee SC, Kim MK et al (2007) T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol 65:453–466. https://doi.org/10.1007/s11103-007-9213-4
Komatsu S, Karibe H, Hamada T, Rakwal R (1999) Phosphorylation upon cold stress in rice (Oryza sativa L.) seedlings. Theor Appl Genet 98:1304–1310
Komatsu S, Yang G, Khan M et al (2007) Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol Gen Genomics 277:713
Koseki M, Kitazawa N, Yonebayashi S et al (2010) Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage. Mol Gen Genomics 284:45–54
Kuk YI, Shin JS, Burgos NR et al (2003) Antioxidative enzymes offer protection from chilling damage in rice plants. Crop Sci 43:2109–2117
Kumar K, Rao KP, Sharma P, Sinha AK (2008) Differential regulation of rice mitogen activated protein kinase kinase (MKK) by abiotic stress. Plant Physiol Biochem 46:891–897. https://doi.org/10.1016/j.plaphy.2008.05.014
Kuroki M, Saito K, Matsuba S et al (2007) A quantitative trait locus for cold tolerance at the booting stage on rice chromosome 8. Theor Appl Genet 115:593–600
Kuroki M, Saito K, Matsuba S (2009) Quantitative trait locus analysis for cold tolerance at the booting stage in a rice cultivar, Hatsushizuku. Jpn Agric Res Q 43:115–121
Kwak TS (1984) Inheritance of seedling cold tolerance in rice. SABRAO J 16:83–86
Lee M-H (2001) Low temperature tolerance in rice: the Korean experience. In: ACIAR Proceedings 1998. ACIAR, Canberra, ACT, pp 109–117
Lee T, Lur H, Chu C (1993) Role of abscisic acid in chilling tolerance of rice (Oryza sativa L.) seedlings. I. Endogenous abscisic acid levels. Plant Cell Environ 16:481–490
Lee DG, Ahsan N, Lee SH et al (2009a) Chilling stress-induced proteomic changes in rice roots. J Plant Physiol 166:1–11
Lee SJ, Jeon US, Lee SJ et al (2009b) Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc Natl Acad Sci U S A 106:22014–22019. https://doi.org/10.4324/9780203605912
Lee J, Lee W, Kwon S-W (2015) A quantitative shotgun proteomics analysis of germinated rice embryos and coleoptiles under low-temperature conditions. Proteome Sci 13:27
Lee J, Heath LS, Grene R, Li S (2019) Comparing time series transcriptome data between plants using a network module finding algorithm. Plant Methods 15:61
Li HB, Wang J, Liu AM et al (1997) Genetic basis of low-temperature-sensitive sterility in indica-japonica hybrids of rice as determined by RFLP analysis. Theor Appl Genet 95:1092–1097
Li G, Zhang M, Cai W-M et al (2008) Characterization of OsPIP2;7, a water channel protein in rice. Plant Cell Physiol 49:1851–1858. https://doi.org/10.1093/pcp/pcn166
Li HW, Zang BS, Deng XW, Wang XP (2011a) Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234:1007–1018. https://doi.org/10.1007/s00425-011-1458-0
Li M, Sun P, Zhou H (2011b) Identification of quantitative trait loci associated with germination using chromosome segment substitution lines of rice (Oryza sativa L.). Theor Appl Genet 123:411–420. https://doi.org/10.1007/s00122-011-1593-9
Li L, Liu X, Xie K et al (2013) qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor Appl Genet 126:2313–2322
Li M, Li X, Zhou Z et al (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 7:377
Li S, Tian Y, Wu K et al (2018) Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560:595. https://doi.org/10.1038/s41586-018-0415-5
Liang Y, Meng L, Lin X et al (2018) QTL and QTL networks for cold tolerance at the reproductive stage detected using selective introgression in rice. PLoS One 13:e0200846
Liu K, Wang L, Xu Y et al (2007) Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta 226:1007–1016. https://doi.org/10.1007/s00425-007-0548-5
Liu C, Wu Y, Wang X (2012) bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235:1157–1169
Liu F, Xu W, Song Q et al (2013) Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol Plant 6:757–767
Lone JA, Khan MN, Bhat MA et al (2018) Cold tolerance at germination and seedling stages of rice: methods of evaluation and characterization of thirty rice genotypes under stress conditions. Int J Curr Microbiol App Sci 7:1103–1109
Lou Q, Chen L, Sun Z et al (2007) A major QTL associated with cold tolerance at seedling stage in rice (Oryza sativa L.). Euphytica 158:87–94. https://doi.org/10.1007/s10681-007-9431-5
Lourenço T, Sapeta H, Figueiredo DD et al (2013) Isolation and characterization of rice (Oryza sativa L.) E3-ubiquitin ligase OsHOS1 gene in the modulation of cold stress response. Plant Mol Biol 83:351–363
Ma Q, Dai X, Xu Y et al (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150:244–256
Ma Y, Dai X, Xu Y et al (2015) COLD1 confers chilling tolerance in rice. Cell 160:1209–1221
Maraña C, García Olmedo F, Carbonero Zalduegui P (1990) Differential expression of two types of sucrose synthase-encoding genes in wheat in response to anaerobiosis, cold shock and light. Gene 88:167–172
Matlon P, Randolph T, Guei R (1998) Impact of rice research in West Africa. In: Impact of rice research. International Rice Research Institute, Los Baños, pp 383–404
Mishra R, Joshi RK, Zhao K (2018) Genome editing in rice: recent advances, challenges, and future implications. Front Plant Sci 9:1361
Miura K, Lin SY, Yano M, Nagamine T (2001) Mapping quantitative trait loci controlling low temperature germinability in rice (Oryza sativa L.). Breed Sci 51:293–299
Mohanty S, Wassmann R, Nelson A et al (2012) Rice and climate change: significance for food security and vulnerability. IRRI discussion paper series No. 49. International Rice Research Institute, Los Baños. 14 p
Molla KA, Yang Y (2019) CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol 37:1121–1142
Moraes De Freitas GP, Basu S, Ramegowda V et al (2016) Comparative analysis of gene expression in response to cold stress in diverse rice genotypes. Biochem Biophys Res Commun 471:253–259. https://doi.org/10.1016/j.bbrc.2016.02.004
Morsy MR, Almutairi AM, Gibbons J et al (2005) The OsLti6 genes encoding low-molecular-weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene 344:171–180
Morsy MR, Jouve L, Hausman J-F et al (2007) Alteration of oxidative and carbohydrate metabolism under abiotic stress in two rice (Oryza sativa L.) genotypes contrasting in chilling tolerance. J Plant Physiol 164:157–167
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci U S A 101:6309–6314
Murai-Hatano M, Kuwagata T, Sakurai J et al (2008) Effect of low root temperature on hydraulic conductivity of rice plants and the possible role of aquaporins. Plant Cell Physiol 49:1294–1305
Nagamine T (1991) Genic control of tolerance to chilling injury at seedling stage in rice, Oryza sativa L. Jpn J Breed 41:35–40
Nagano K (1998) Development of new breeding techniques for cold tolerance and breeding of new rice cultivars with highly cold tolerance, Hitomebore and Jyoudeki. In: Proceedings of the International Workshop on Breeding and Biotechnology for Environmental Stress in Rice, pp 143–148
Nakashima K, Tran LP, Van Nguyen D et al (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Nalley L, Tack J, Durand A et al (2017) The production, consumption, and environmental impacts of rice hybridization in the United States. Agron J 109:193–203. https://doi.org/10.2134/agronj2016.05.0281
Novillo F, Alonso JM, Ecker JR, Salinas J (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci U S A 101:3985–3990
Oh SJ, Kim YS, Kwon CW et al (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150:1368–1379
Okamoto E, Matsunaga K, Sasaki T (1986) Cold tolerant rice varieties in temperate district of Japan. Tohoku Agric Res 39:35
Oliver SN, Van Dongen JT, Alfred SC et al (2005) Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ 28:1534–1551
Oliver SN, Zhao X, Dennis ES, Dolferus R (2007) The molecular basis of cold-induced pollen sterility in rice. In: Biotechnology and sustainable agriculture 2006 and beyond. Springer, New York, NY, pp 205–207
Pan Y, Zhang H, Zhang D et al (2015) Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS One 10:e0120590. https://doi.org/10.1371/journal.pone.0120590
Pan Y, Liang H, Gao L et al (2020) Transcriptomic profiling of germinating seeds under cold stress and characterization of the cold-tolerant gene LTG5 in rice. BMC Plant Biol 20:1–17
Pang Y, Chen K, Wang X et al (2017) Simultaneous improvement and genetic dissection of salt tolerance of rice (Oryza sativa L.) by designed QTL pyramiding. Front Plant Sci 8:1275. https://doi.org/10.3389/fpls.2017.01275
Park H, Soon I, Han J et al (2009) OsDEG10 encoding a small RNA-binding protein is involved in abiotic stress signaling. Biochem Biophys Res Commun 380:597–602. https://doi.org/10.1016/j.bbrc.2009.01.131
Park M, Yun K, Mohanty B et al (2010) Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ 33:2209–2230
Park IK, Oh CS, Kim DM et al (2013) QTL mapping for cold tolerance at the seedling stage using introgression lines derived from an intersubspecific cross in rice. Plant Breed Biotech 1:1–8. https://doi.org/10.9787/PBB.2013.1.1.001
Pouramir Dashtmian F, Khajeh Hosseini M, Esfahani M (2013) Methods for rice genotypes cold tolerance evaluation at germination stage. Int J Agric Crop Sci 5:2111
Ranawake AL, Nakamura C (2011) Cold tolerance of an inbred line population of rice (Oryza sativa L) at different growth stages. Trop Agric Res Ext 14:25–33
Ranawake A, Nakamura C (2015) QTL analysis of dehydration tolerance at seedling stage in rice (Oryza sativa L.). Trop Agric Res 15:98
Ranawake AL, Manangkil OE, Yoshida S et al (2014) Mapping QTLs for cold tolerance at germination and the early seedling stage in rice (Oryza sativa L.). Biotechnol Biotechnol Equip 28:989–998. https://doi.org/10.1080/13102818.2014.978539
Saad RB, Fabre D, Mieulet D et al (2012) Expression of the Aeluropus littoralis AlSAP gene in rice confers broad tolerance to abiotic stresses through maintenance of photosynthesis. Plant Cell Environ 35:626–643
Saito K, Miura K, Nagano K et al (1995) Chromosomal location of quantitative trait loci for cool tolerance at the booting stage in rice [Oryza sativa] variety ‘Norin-PL8’. Breed Sci 45:337–340
Saito K, Miura K, Nagano K et al (2001) Identification of two closely linked quantitative trait loci for cold tolerance on chromosome 4 of rice and their association with anther length. Theor Appl Genet 103:862–868
Saito K, Hayano-Saito Y, Maruyama-Funatsuki W et al (2004) Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor Appl Genet 109:515–522. https://doi.org/10.1007/s00122-004-1667-z
Sakamoto A, Murata N (2002) The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant Cell Environ 25:163–171
Sales E, Viruel J, Domingo C, Marqués L (2017) Genome wide association analysis of cold tolerance at germination in temperate japonica rice (Oryza sativa L.) varieties. PLoS One 12:e0183416
Sanchez-Ballesta MT, Lafuente MT, Zacarias L, Granell A (2000) Involvement of phenylalanine ammonia-lyase in the response of Fortune mandarin fruits to cold temperature. Physiol Plant 108:382–389
Satake T (1976) Sterile-type cool injury in paddy rice plants. In: Proceedings of the Symposium on Climate and Rice. International Rice Research Institute (IRRI), Los Baños, pp 281–300
Sato Y, Masuta Y, Saito K et al (2011) Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep 30:399–406
Satoh T, Tezuka K, Kawamoto T et al (2016) Identification of QTLs controlling low-temperature germination of the East European rice (Oryza sativa L.) variety Maratteli. Euphytica 207:245–254. https://doi.org/10.1007/s10681-015-1531-z
Schiller JM, Linquist B, Douangsila K et al (2001) Constraints to rice production systems in Laos. In: ACIAR Proceedings 1998. ACIAR, Canberra, ACT, pp 3–19
Schläppi MR, Jackson AK, Eizenga GC et al (2017) Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA Mini-Core collection. Front Plant Sci 8:957. https://doi.org/10.3389/fpls.2017.00957
Seck PA, Diagne A, Mohanty S, Wopereis MCS (2012) Crops that feed the world 7: rice. Food Secur 4:7–24
Serrano-Mislata A, Bencivenga S, Bush M et al (2017) DELLA genes restrict inflorescence meristem function independently of plant height. Nat Plants 3:749
Shakiba E, Edwards JD, Jodari F et al (2017) Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS One 12:1–22. https://doi.org/10.1371/journal.pone.0172133
Sharma KD, Nayyar H (2016) Regulatory networks in pollen development under cold stress. Front Plant Sci 7:402
Shi J, Cao Y, Fan X (2012) A rice microsomal delta-12 fatty acid desaturase can enhance resistance to cold stress in yeast and Oryza sativa. Mol Breed 29:743–757. https://doi.org/10.1007/s11032-011-9587-5
Shim K-C, Kim S, Le AQ et al (2019) Fine mapping of a low-temperature germinability QTL qLTG1 using introgression lines derived from Oryza rufipogon. Plant Breed Biotechnol 7:141–150
Shinada H, Iwata N, Sato T, Fujino K (2013) Genetical and morphological characterization of cold tolerance at fertilization stage in rice. Breed Sci 63:197–204. https://doi.org/10.1270/jsbbs.63.197
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223
Shirasawa K, Shiokai S, Yamaguchi M et al (2006) Dot-blot-SNP analysis for practical plant breeding and cultivar identification in rice. Theor Appl Genet 113:147–155
Shirasawa S, Endo T, Nakagomi K et al (2012) Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar,‘Lijiangxintuanheigu’, in rice, Oryza sativa L. Theor Appl Genet 124:937–946
Singh BK, Sutradhar M, Singh AK, Mandal N (2017) Cold stress in rice at early growth stage: an overview. Int J Pure Appl Biosci 5:407–419
Sinha SK, Amitha Mithra SV, Chaudhary S et al (2018) Transcriptome analysis of two rice varieties contrasting for nitrogen use efficiency under chronic N starvation reveals differences in chloroplast and starch metabolism-related genes. Genes (Basel) 9:1–22. https://doi.org/10.3390/genes9040206
Sivapalan S (2013) Cold tolerance of temperate and tropical rice varieties. Frank Wise Institute of Tropical Agriculture, Kununurra, WA. 4 p
Snell P, Johnston D, Ford R (2008) Cold tolerant rice varieties: a matter of need for Australia. IREC Farm Newsl 177:4–5
Song S-Y, Chen Y, Chen J et al (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345
Song J, Li J, Sun J et al (2018) Genome-wide association mapping for cold tolerance in a core collection of rice (Oryza sativa L.) landraces by using high-density single nucleotide polymorphism markers from specific-locus amplified fragment sequencing. Front Plant Sci 9:875
Sthapit BR, Witcombe JR (1998) Inheritance of tolerance to chilling stress in rice during germination and plumule greening. Crop Sci 38:660–665
Su CF, Wang YC, Hsieh TH et al (2010) A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol 153:145–158. https://doi.org/10.1104/pp.110.153015
Suh JP, Jeung JU, Lee JI et al (2010) Identification and analysis of QTLs controlling cold tolerance at the reproductive stage and validation of effective QTLs in cold-tolerant genotypes of rice (Oryza sativa L.). Theor Appl Genet 120:985–995
Suh JP, Lee CK, Lee JH et al (2012) Identification of quantitative trait loci for seedling cold tolerance using RILs derived from a cross between japonica and tropical japonica rice cultivars. Euphytica 184:101–108
Sun Z, Du J, Pu X et al (2019) Near-isogenic lines of japonica rice revealed new QTLs for cold tolerance at booting stage. Agronomy 9(1):40. https://doi.org/10.3390/agronomy9010040
Sweeney M, McCouch S (2007) The complex history of the domestication of rice. Ann Bot 100:951–957
Takahashi N (1984) Differentiation of ecotypes in Oryza sativa L. In: Developments in crop science. Elsevier, Amsterdam, pp 31–67
Takesawa T, Ito M, Kanzaki H et al (2002) Over-expression of ζ glutathione S-transferase in transgenic rice enhances germination and growth at low temperature. Mol Breed 9:93–101
Tao Z, Kou Y, Liu H et al (2011) OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62:4863–4874. https://doi.org/10.1093/jxb/err144
Teng S, Zeng D, Qian Q et al (2001) QTL analysis of rice low temperature germinability. Chin Sci Bull 46:1800–1803
Urano K, Maruyama K, Ogata Y et al (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57:1065–1078
Vats S, Kumawat S, Kumar V et al (2019) Genome editing in plants: exploration of technological advancements and challenges. Cell 8:1386
Wainaina CM, Makihara D, Nakamura M et al (2018) Identification and validation of QTLs for cold tolerance at the booting stage and other agronomic traits in a rice cross of a Japanese tolerant variety, Hananomai, and a NERICA parent, WAB56-104. Plant Prod Sci 21:132–143
Wang Q, Guan Y, Wu Y et al (2008) Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol 67:589–602
Wang ZF, Wang JF, Wang FH et al (2009) Genetic control of germination ability under cold stress in rice. Rice Sci 16:173–180. https://doi.org/10.1016/S1672-6308(08)60076-1
Wang Z, Wang F, Zhou R et al (2011) Identification of quantitative trait loci for cold tolerance during the germination and seedling stages in rice (Oryza sativa L.). Euphytica 181:405
Wang C, Wei Q, Zhang K et al (2013a) Down-regulation of OsSPX1 causes high sensitivity to cold and oxidative stresses in rice seedlings. PLoS One 8(12):e81849. https://doi.org/10.1371/journal.pone.0081849
Wang J, Lin X, Sun Q, Jena KK (2013b) Evaluation of cold tolerance for japonica rice varieties from different countries. Adv J Food Sci Technol 5:54–56. https://doi.org/10.19026/ajfst.5.3311
Wang X, Han F, Yang M et al (2013c) Exploring the response of rice (Oryza sativa) leaf to gibberellins: a proteomic strategy. Rice 6:17
Wang D, Liu J, Li C et al (2016) Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice 9:61. https://doi.org/10.1186/s12284-016-0133-2
Wei J, Wang A, Li R et al (2018) Metabolome-wide association studies for agronomic traits of rice. Heredity (Edinb) 120:342
Williams ME, Torabinejad J, Cohick E et al (2005) Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol 138:686–700
Wittmann-Liebold B, Graack H, Pohl T (2006) Two-dimensional gel electrophoresis as tool for proteomics studies in combination with protein identification by mass spectrometry. Proteomics 6:4688–4703
Wolter F, Schindele P, Puchta H (2019) Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol 19:176
Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144:1416–1428. https://doi.org/10.1104/pp.107.101295
Xiao N, Huang W, Li A et al (2015) Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Theor Appl Genet 128:173–185
Xiao N, Gao Y, Qian H et al (2018) Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol 177:1108–1123
Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid–inducible mitogen-activated protein kinase. Plant Cell 15:745–759
Xu L-M, Zhou L, Zeng Y-W et al (2008) Identification and mapping of quantitative trait loci for cold tolerance at the booting stage in a japonica rice near-isogenic line. Plant Sci 174:340–347
Xu R, Yang Y, Qin R et al (2016) Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J Genet Genom 43:529–532
Xu Y, Hu D, Hou X et al (2020) OsTMF attenuates cold tolerance by affecting cell wall properties in rice. New Phytol 227:498. https://doi.org/10.1111/nph.16549
Yamaguchi-Shinozaki K, Shinozaki K (2004) Improving abiotic stress tolerance in crops. In The 8th International Symposium on the Biosafety of Genetically Modified Organisms, Montpellier, France, 26–30 September, 2004, International Society for Biosafety Research, pp 134–136
Yamori W, Noguchi KO, Hikosaka K, Terashima I (2010) Phenotypic plasticity in photosynthetic temperature acclimation among crop species with different cold tolerances. Plant Physiol 152:388–399
Yamori W, Sakata N, Suzuki Y et al (2011) Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant J 68:966–976. https://doi.org/10.1111/j.1365-313X.2011.04747.x
Yan SP, Zhang QY, Tang ZC et al (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics 5:484–496
Yang T, Poovaiah BW (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8:505–512
Yang G, Rhodes D, Joly RJ (1996) Effects of high temperature on membrane stability and chlorophyll fluorescence in glycinebetaine-deficient and glycinebetaine-containing maize lines. Funct Plant Biol 23:437–443
Yang T, Shad Ali G, Yang L et al (2010) Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal Behav 5:991–994
Yang A, Dai X, Zhang W-H (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556
Yang C, Li D, Mao D et al (2013a) Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ 36:2207–2218
Yang Z, Huang D, Tang W et al (2013b) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One 8:e68433
Yang W, Guo Z, Huang C et al (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5:5087
Yang LM, Liu HL, Lei L et al (2018) Identification of QTLs controlling low-temperature germinability and cold tolerance at the seedling stage in rice (Oryza sativa L.). Euphytica 214:13. https://doi.org/10.1007/s10681-017-2092-0
Ye C, Fukai S, Godwin I et al (2009) Cold tolerance in rice varieties at different growth stages. Crop Pasture Sci 60:328–338
Yokotani N, Sato Y, Tanabe S et al (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64:5085–5097. https://doi.org/10.1093/jxb/ert298
Yoshida S (1981) Fundamentals of rice crop science. International Rice Research Institute, Los Baños
Yu S, Li M, Xiao Y et al (2018) Mapping QTLs for cold tolerance at seedling stage using an Oryza sativa × O. rufipogon backcross inbred line population. Czech J Genet Plant Breed 54:59–64. https://doi.org/10.17221/154/2016-CJGPB
Yuan P, Yang T, Poovaiah BW (2018) Calcium signaling-mediated plant response to cold stress. Int J Mol Sci 19:3896
Yun KY, Park MR, Mohanty B et al (2010) Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol 10:16
Zhang X, Fowler SG, Cheng H et al (2004) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J 39:905–919
Zhang Y, Chen C, Jin XF et al (2009) Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep 42:486–492
Zhang J, Li J, Wang X, Chen J (2011) OVP1, a vacuolar H+-translocating inorganic pyrophosphatase (V-PPase), overexpression improved rice cold tolerance. Plant Physiol Biochem 49:33–38
Zhang T, Zhao X, Wang W et al (2012a) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7:e43274
Zhang Y, Xu Y, Yi H, Gong J (2012b) Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J 72:400–410
Zhang K, Liu H, Tao P, Chen H (2014a) Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. PLoS One 9:e98215. https://doi.org/10.1371/journal.pone.0098215
Zhang Q, Chen Q, Wang S et al (2014b) Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 7:24. https://doi.org/10.1186/s12284-014-0024-3
Zhang S, Zheng J, Liu B et al (2014c) Identification of QTLs for cold tolerance at seedling stage in rice (Oryza sativa L.) using two distinct methods of cold treatment. Euphytica 195:95–104
Zhang J, Luo W, Zhao Y et al (2016) Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice. New Phytol 211:1295–1310
Zhang Z, Li JJJ, Pan Y et al (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat Commun 8:1–13. https://doi.org/10.1007/BF01337500
Zhang M, Ye J, Xu Q et al (2018) Genome-wide association study of cold tolerance of Chinese indica rice varieties at the bud burst stage. Plant Cell Rep 37:529–539
Zhao WG, Chung JW, Kwon SW et al (2013) Association analysis of physicochemical traits on eating quality in rice (Oryza sativa L.). Euphytica 191:9–21
Zhi-Hong Z, Li S, Wei L et al (2005) A major QTL conferring cold tolerance at the early seedling stage using recombinant inbred lines of rice (Oryza sativa L.). Plant Sci 168:527–534. https://doi.org/10.1016/j.plantsci.2004.09.021
Zhou L, Zeng Y, Hu G et al (2012) Characterization and identification of cold tolerant near-isogenic lines in rice. Breed Sci 62:196–201
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324
Zhu Y, Chen K, Mi X et al (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS One 10:e0145704. https://doi.org/10.1371/journal.pone.0145704
Acknowledgments
The authors would like to thank and acknowledge the Bill & Melinda Gates Foundation for providing a research grant to ZL for the Green Super Rice project under ID OPP1130530. We would also like to thank the Department of Agriculture, Philippines, for providing funds to JA under the Next-Gen project, and also thank and acknowledge IRRI Communications for English language editing and anonymous internal reviewers for their valuable suggestions and constructive comments that helped improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this chapter
Cite this chapter
Najeeb, S., Mahender, A., Anandan, A., Hussain, W., Li, Z., Ali, J. (2021). Genetics and Breeding of Low-Temperature Stress Tolerance in Rice. In: Ali, J., Wani, S.H. (eds) Rice Improvement. Springer, Cham. https://doi.org/10.1007/978-3-030-66530-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-66530-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66529-6
Online ISBN: 978-3-030-66530-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)