Abstract
Key message
A region containing three genes on chromosome 1 of indica rice was associated with cold tolerance at the bud burst stage; these results may be useful for breeding cold-tolerant lines.
Abstract
Low temperature at the bud burst stage is one of the major abiotic stresses limiting rice growth, especially in regions where rice seeds are sown directly. In this study, we investigated cold tolerance of rice at the bud burst stage and conducted a genome-wide association study (GWAS) based on the 5K rice array of 249 indica rice varieties widely distributed in China. We improved the method to assess cold tolerance at the bud burst stage in indica rice, and used severity of damage (SD) and seed survival rate (SR) as the cold-tolerant indices. Population structure analysis demonstrated that the Chinese indica panel was divided into three subgroups. In total, 47 significant single-nucleotide polymorphism (SNP) loci associated with SD and SR, were detected by association mapping based on mixed linear model. Because some loci overlapped between SD and SR, the loci contained 13 genome intervals and most of them have been reported previously. A major QTL for cold tolerance on chromosome 1 at the position of 31.6 Mb, explaining 13.2% of phenotypic variation, was selected for further analysis. Through LD decay, GO enrichment, RNA-seq data, and gene expression pattern analyses, we identified three genes (LOC_Os01g55510, LOC_Os01g55350 and LOC_Os01g55560) that were differentially expressed between cold-tolerant and cold-sensitive varieties, suggesting they may be candidate genes for cold tolerance. Together, our results provide a new method to assess cold tolerance in indica rice, and establish the foundation for isolating genes related to cold tolerance that could be used in rice breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is one of the most important staple crops, feeding more than 60% of the population in China and providing 21% of energy per capita on a global scale (Cheng et al. 2007). Unlike other cereals such as wheat and barley, rice is a temperature-sensitive crop that can be injured by exposure to low temperatures (Sipaseuth et al. 2007). Low temperature is one of the most common environmental stress factors; it can affect rice architecture and seed germination, slow the growth rate, delay seed maturation, cause male sterility, decrease the seed setting rate, and ultimately reduce the rice yield (Fujino and Matsuda 2010; Fujino et al. 2008; Ma et al. 2015; Saito et al. 2004, 2010; Suh et al. 2010). Cold stress is a major factor contributing to reduced rice yield in temperate and high-altitude regions. In the world, about 15 million hectares of land are prone to cold temperatures, especially in Japan, Korea, and Northeast and Southwest China (Sthapit and Witcombe 1998). During the planting season, when the climate can be unpredictable, pre-germinated rice seeds planted directly in soil can be affected by low temperatures of the air and/or irrigation water, which can greatly decrease the germination rate and even cause seed death (Fujino and Matsuda 2010). Additionally, cold stress at the reproductive stage can negatively affect grain quality and production (Lee 2001; Xu et al. 2008). Therefore, improving the cold tolerance of rice is an important objective in rice breeding, not only to maintain rice yields in cool regions, but also to expand the cultivation area of rice into northern areas or high-altitude regions with low temperatures (Ma et al. 2015). Consequently, cold-tolerant rice cultivars would benefit grain production and contribute to food security and continuing development.
In rice, cold tolerance is a very complex quantitative trait that is genetically controlled by multiple quantitative trait loci (QTLs). One of the most common methods to study the genetic basis of cold tolerance in rice is QTL analysis. Many genomic regions on all 12 rice chromosomes have been reported to contain QTLs for cold tolerance at different development stages. Most of these QTLs have been detected repeatedly using different bi-parental populations. Generally, japonica cultivars are more cold-tolerant than indica cultivars (Shakiba et al. 2017). Most bi-parental populations used in QTL analyses have been derived from a cross between a cold-tolerant japonica variety and a cold-sensitive indica variety; consequently, most QTLs associated with cold tolerance are derived from the japonica parent (Andaya and Mackill 2003; Kuroki et al. 2007; Ma et al. 2015; Zhu et al. 2015). From these QTLs, several genes have been cloned using recombinant inbred lines (RILs) and backcross inbred line (BILs) (Ma et al. 2015; Saito et al. 2010; Zhang et al. 2014). Among the genes cloned so far, LTG3-1 is related to tolerance at the germination stage; this gene encodes a protein of unknown function that may be involved in tissue weakening (Fujino et al. 2008). Ctb1, which was cloned from a QTL for cold tolerance at the booting stage of rice, encodes an F-box protein that interacts with a subunit of the E3 ubiquitin ligase, Skp1. This suggests that an ubiquitin–proteasome pathway is involved in cold tolerance at the booting stage (Saito et al. 2010). COLD1 encodes a regulator of G-protein signaling, conferring cold tolerance at the seedling stage in japonica rice (Ma et al. 2015). qCTS-9 is the latest gene to be identified, and encodes unknown expressed protein, contributing to enhance cold tolerance at the seedling stage in rice (Zhao et al. 2017). To better understand the genetic mechanism of cold tolerance, more genes related to this trait should be identified. Although bi-parental mapping populations are a good method for gene cloning, limitation of genetic diversity and long time period with less resolution are the shortcoming for discovering more new genes (Pradhan et al. 2016). The genome-wide association study (GWAS) method has proved to be very useful for dissecting complicated quantitative traits based on a linkage disequilibrium mapping approach (Huang et al. 2010, 2012b, 2016). Recently, several studies have used GWAS to explore cold tolerance in rice. 17 QTLs related to rice germinability at low temperature were detected by a GWAS analysis with the population of 63 rice varieties from Japan (Fujino et al. 2015). Pan et al. (2015) mapped 51 QTLs for cold tolerance at the germination and booting stages using a population of 174 Chinese rice varieties. Lv et al. (2016) detected 132 QTL for nature chilling and cold-shock tolerance at the seedling stage by a GWAS analysis of 527 rice cultivars. Wang et al. (2016) identified 67 QTLs for cold tolerance at the seedling stage, and 56 of these QTLs were located in regions that had not been reported to contain cold tolerance-related QTLs. Shakiba et al. (2017) identified 42 QTLs associated with cold tolerance at the seedling stage, 20 of which did not co-localize with previously reported cold-tolerant QTLs. Consequently, GWAS can identify new QTLs for cold tolerance and provide new insights into the genetic basis of cold tolerance in rice.
To study the genetic mechanism of cold tolerance in rice, the methods evaluating the cold tolerance is the key point. Cold stress has different effects at different stages. At the germination stage, the germination vigor and seedling survival rate are the two main criteria used to evaluate cold tolerance (Han et al. 2006; Zhou et al. 2012). At the seedling stage, degree of cold tolerance can be evaluated by scoring seedling injury and chlorosis (Nagamine 1991). Additionally, survival percentage is another widely used criterion to assess cold tolerance at the seedling stage (Lv et al. 2016; Ma et al. 2015; Schlappi et al. 2017). At the reproductive stage, changes in the seed setting rate or individual yield after low temperature treatment are also commonly used to evaluate cold tolerance (Shirasawa et al. 2012). At the bud burst stage, similar to the seedling stage, low temperature can affect the seed shoot growth and seed survival rate. Generally, seed survival rate is widely used to evaluate cold tolerance. Cold-tolerant seeds can grow normally and became a seedling after low temperature treatment, while cold-sensitive seeds grow slowly or almost died. With the increasing of the labor cost and the improvement of mechanization, sowing the germinated rice seeds directly in the field became popular in China. Cold tolerance at the bud burst stage became more and more important trait in rice breeding, but its genetic basic is still poorly understood even in the indica population.
In this study, 249 indica varieties widely distributed in China were collected to evaluate cold tolerance at the bud burst stage using two different criteria, and were genotyped with a 5K SNP array. Then, GWAS analysis was performed to identify new QTLs for cold tolerance in indica rice at the bud burst stage. These results will be useful for improving cold tolerance in rice breeding and for discovering new genes related to cold tolerance.
Materials and methods
Plant material
The natural population comprised 249 indica rice varieties, which were collected from China (Table S2). These varieties were chosen from the previous study (Xu et al. 2016), so as to cover the largest geographical region planted with indica rice in southern China. They were grown for more than two generations (planting density, 20 cm × 20 cm) at the Experimental Farm of the China National Rice Research Institute to ensure homogeneity, and their seeds were obtained from the middle plants in each line.
Cold tolerance evaluation at the bud burst stage
Rice seeds were air-dried naturally, and kept at 55 °C for 5 days to break dormancy. Then, the seeds were surface-sterilized with 70% ethyl alcohol and washed three times with sterile water. Next, the seeds were soaked in water for about 2 days and allowed to germinate for 1 day. Thirty seeds with 5-mm long shoots were selected, washed with sterile water, and transferred onto wet absorbent filter paper in a culture dish. The germinated seeds were then subjected to a cold treatment at 5 °C for 5 days in a growth chamber in darkness. After that, severity of damage (SD) was assessed after 3 days of recovery growth in a new chamber. The SD was scored as follows: Score 0, the seedling had normal leaf color and grew well with no damage; Score 1, the seed grew well with little damage and the leaf color was green; Score 3, the shoot grew slowly and the shoot color was green; Score 5, the shoot size had not increased and the internal leaf color was green, but the cotyledon was withered; Score 7, the germinated seed was dead with no green leaves. The SD value of one variety represented the average scores of 30 seeds. The conditions in the chamber for recovery growth were as follows: 16-h light/8-dark photoperiod (32 °C/28 °C), and relative humidity of 70%. The seed survival rate (SR) was evaluated after 6 days of recovery growth, and was calculated as follows: seedling survival rate (%) = surviving seedlings/30 × 100 (Zhou et al. 2012). All experiments were performed in triplicate.
Genotyping and population structure analysis
All 249 indica rice varieties were genotyped by the 5K SNPs rice array. We used a subset of 3867 SNP markers selected from the whole SNP array by applying the following thresholds: missing data ratio > 90% and markers with frequency of minor allele (MAF) > 0.05. A neighbor-joining (NJ) tree was constructed using MEGA6 software (Tamura et al. 2013) based on Nei’s genetic distance between pairwise individuals under PowerMarker version 3.25 (Liu and Muse 2005). EIGENSOFT software was used to conduct a principal components analysis (PCA) to estimate the number of subpopulations (Patterson et al. 2006). ADMIXTURE software was used to calculate the genetic component for each variety (Alexander et al. 2009).
GWAS analysis of cold tolerance at the bud burst stage
The GWAS analysis was performed with a line mixed-effects model to determine the association between genotype and evaluated phenotype using EMMAX software (Kang et al. 2010). The kinship matrix was calculated by EMMAX-kin to measure the genetic similarities between individuals. Three PCs were selected as population structure to correct the GWAS results. To obtain independent association signals, multiple SNPs passing the threshold on the same chromosome were clustered as one association locus, and the SNP with the minimum P value in a cluster was considered as the lead SNP.
RNA extraction and quantitative real-time PCR analysis
Five shoots from each variety were sampled under cold and normal conditions, respectively. Total RNA was extracted from rice shoot using a MiniBEST Plant RNA Extraction kit (Takara Bio Inc., Otsu, Japan). All samples were treated with DNase I (Takara Bio Inc.). Complementary DNA was synthesized from total RNA using PrimeScript RT Master Mix (Takara Bio Inc.). Quantitative real-time PCR (qRT-PCR) was performed using a 2 × SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) on an Applied Biosystems 7500 Real-time PCR System. Table S1 summarizes the gene accessions and primers used for qRT-PCR in this study. The mRNA level of these genes was determined with the housekeeping gene Actin as an internal control. Data shown in figures and tables are mean values of three repeats.
Statistical analysis
Kolmogorov–Smirnov tests, correlation analyses, and Duncan’s multiple comparisons were performed using SAS 8.0 (SAS, Inc., Cary, NC, USA).
Results
Geographical distribution of rice varieties
There were wide variations in cold tolerance at the bud burst stage among the rice germplasm resource (Fig. 1b). Here, the 249 indica rice varieties including 165 old landraces and 84 improved varieties were selected from different provinces of China (Fig. 1a, Table S2). They were distributed in 16 provinces or municipalities covering all the regions southern China. Most of the rice varieties were planted before the 1970s in China; these landraces have higher genetic diversity than current breeding varieties (Xu et al. 2016). The panel had a wide representation of Chinese indica varieties and was suitable for GWAS analysis.
Geographical distribution and germplasm diversity of cold tolerance. a Geographical distribution of indica rice varieties. b Variations in cold tolerance at bud burst stage in rice germplasm. Bar = 5 cm. The dish at the top left showed the most in-tolerant to cold stress, and the one at the bottom right showed the most tolerant to cold stress
Variation in cold tolerance among the 249 indica rice varieties
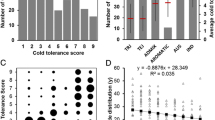
In our study, two cold tolerance indices at the bud burst stage were evaluated: SD and SR (Table S3). Because indica rice varieties are very cold-sensitive, a mild cold stress treatment was used in this study. The germinated seeds were exposed to low temperature for only 5 days and SD was scored after 3 days of recovery growth. This method differed from that in a previous report (Han and Zhang 2004). According to the phenotype of the germinated seeds (shoot color, growth rate, growth vigor, and chlorosis) each seed was scored with a resistance level (score 0, 1, 3, 5, or 7) (Fig. 2a). The average of all seeds’ scores was the score for that variety. Another cold tolerance index, SR, was also evaluated after 6 days of recovery growth. Interestingly, almost all the seeds with scores 0 and 1 survived, while almost all with scores of 5 and 7 died, but seeds with score 3 contained both two results above (Fig. 2b). The correlation analysis suggested that there was a strong relationship between SD and SR, with the correlation coefficient was − 0.8878.
The scores of SD and SR showed continuous distributions in the indica rice panel (Fig. 3a, c). However, the SD scores were concentrated around 5–6, and the SR scores were focused on 0–20% (Fig. 3b, d). It was also found that the phenotypic segregation of SD was closer to fitting a normal distribution, as indicated by the Kolmogorov–Smirnov test (D = 0.15) than was SR (D = 0.22). Therefore, the cold tolerance index of SD would be more suitable for GWAS mapping than SR in this indica population.
Population structure and relative kinship
Based on the nucleotide polymorphisms, we calculated the genetic component of each variety using admixture software. The one with the lowest cross-validation (CV) error was selected to evaluate the number of subgroups. As a result, K = 3 was selected, suggesting that the indica panel could be divided into three subgroups (I, II, and III) (Fig. 4a, Table S4). The PCA demonstrated that the indica panel formed three subgroups with different distributions along the two eigenvectors; PC1 and PC2 accounted for 9.7 and 5.3% of the genetic variation, respectively (Fig. 4b). Additionally, a NJ tree was constructed based on Nei’s genetic distances with the three clusters (blue, orange, and grey) (Fig. 4c). The combined results of the NJ tree and the PCA indicated that, although the indica panel could be divided into three subgroups, these indica varieties did not show a strong population structure. In the pairwise relative kinship values analysis, more than half of the kinship coefficient values were around zero and 81% of the all values were less than 0.1 (Fig. 4d). Less than 1% of the values were larger than 0.5. Together, these results indicated that there was weak relatedness among our indica population, which was beneficial for subsequent GWAS mapping.
Population structure of 249 indica rice. a Subgroups (K = 3) inferred using admixture software; b principal component analysis of rice panel; c neighbor-joining tree based on Nei’s genetic distances; d pairwise relative kinship analysis of rice panel. Blue, orange, and grey represent subgroup I, II, and III, respectively
GWAS analysis for cold tolerance at the bud burst stage
Association mapping was performed under a mixed linear model with the PCA matrix (Table S5) and kinship matrix as covariates. In total, 47 significant SNP loci associated with SR and SD were detected at the threshold of 2.5 across all 12 chromosomes with well-fitted quantile–quantile (Q–Q) plots (Fig. 5). Comparison between the GWAS results for SR and SD revealed that most of the significant SNPs were detected in both SR and SD, indicating that these two indexes were strongly connected. In the GWAS for SR, 26 SNP loci distributed on chromosomes 1, 3, 4, 5, 6, 10, and 12 were detected containing 12 genome intervals, named qCTSR1-1, qCTSR1-2, qCTSR1-3, qCTSR3-1, qCTSR3-2, qCTSR4-1, qCTSR5-1, qCTSR5-2, qCTSR6-1, qCTSR10-1, qCTSR12-1 and qCTSR12-2, respectively (Table 1). All of them except qCTSR1-3 and qCTSR3-2 were also detected in the GWAS for SD. Moreover, qCTSD11-1 was only detected in the GWAS for SD. Among these QTLs, qCTSR1-2 overlapped with qCTSD1-2, which harbored the highest-peak SNP, rs328, which explain 13.2% of the total phenotypic variation. The QTLs were located on chromosome 1 near the position 31.6 Mb with the lowest P value. Interestingly, 7 of the 10 common QTLs had a higher – log10(P) value in the GWAS for SR than in the GWAS for SD (Table 1, Fig. S1). This may be because the survival rate amplified the difference among varieties. For example, two varieties with the similar score may exhibit different survival status. Likewise, varieties with same survival status can have different SD scores. Therefore, there were still some QTLs with lower – log10(P) values in the GWAS for SR.
Manhattan plots and quantile–quantile (Q–Q) plot of GWAS for SD and SR. Manhattan plot of GWAS for SD (a) and SR (c). Blue line indicates threshold of P value; Q–Q plot of GWAS for SD b and SR d, red straight line represents expected null distribution of P values, black dots represent observed distribution of P values. λ represented the value of inflation factor, calculated by R software (http://www.r-project.org/)
We compared the significant SNP loci detected in this study with QTLs detected previously by linkage or association mapping. There were many overlaps between our QTLs and those previously reported to be associated with cold tolerance (Table 1). Among these co-localized regions, there was no known functional gene, such as COLD1, qLTG3-1, or Ctb1. Only three QTLs for SR detected in our study have not been reported previously. Two of them, named qCTSR1-3 and qCTSR3-2, were not detected in the GWAS for SD. Additionally, except for qCTSD12-1, all of the QTLs for SD have been reported previously. Specifically, qCTSR1-2 and qCTSR5-2 detected in our study were also identified in a bulked segregant analysis (BSA) using high-throughput sequencing of pooled extremes (Yang et al. 2013). qCTSR5-1 and qCTSR10-1 were detected in a previous study by linkage mapping using bi-parents and their derived population under low-temperature stress (Jiang et al. 2006). Liu et al. (2013) identified a QTL for cold stress named qCTS6, which overlapped with qCTSR6-1. Furthermore, qCTSR1-1, qCTSR3-1, qCTSR4-1, and qCTSR12-2 were also detected by Lv et al. (2016) using association mapping, and overlapped with the loci L7, L27, L43, and L131, respectively. Together, these results verified the accuracy of our GWAS study.
Candidate gene analysis
The GWAS analysis revealed a highly significant site for cold tolerance on chromosome 1 at about 31.6 Mb (Fig. 6a). According to the LD decay analysis, a total 579-kb region was identified as the candidate region (Fig. 6b), which contained 89 genes including 48 functionally annotated genes, 29 expressed proteins with unknown function, and 11 retro-transposon proteins (Table S6). According to the gene function annotation and GO enrichment analysis (Table S7), we chose genes with functions related to stress response or metabolic process. We also referred to RNA-seq data reported previously (Shen et al. 2014). As a result, we selected 18 candidate genes to compare expression levels between cold-tolerant and cold-sensitive varieties by qRT-PCR analysis. In these analyses, three genes (LOC_Os01g55510, LOC_Os01g55350 and LOC_Os01g55560) were differentially expressed between four cold-tolerant varieties and four cold-sensitive varieties (Fig. 7). LOC_Os01g55510 encodes dynein light chain type 1 domain containing protein, its homologous protein in Arabidopsis participates in the microtubule-based process. LOC_Os01g55350 encodes phosphoenolpyruvate carboxylase, which was the key enzyme in the last step of glycolytic pathway and expressed in response to abiotic stress (Sánchez et al. 2006). LOC_Os01g55560 encodes ABIL3 protein, which is the subunits of SCAR/WAVE complex associated with microtubule cytoskeleton (Jörgens et al. 2010). These genes showed higher transcript levels in cold-tolerant varieties than in cold-sensitive varieties under normal growth conditions. Under cold stress conditions, similar expression patterns were observed. Among these three genes, two (LOC_Os01g55350 and LOC_Os01g55560) showed higher transcript levels in cold-tolerant varieties under cold stress conditions than under normal growth conditions, but no change in cold-sensitive varieties between the two conditions. The transcript level of LOC_Os01g55510 in both cold-sensitive and cold-tolerant varieties were up-regulated under cold stress conditions. However, the other 15 genes did not display any differential expression between cold-tolerant and cold-sensitive varieties under normal growth conditions and cold stress conditions (Table S8). Further experiments including genetic complementation analyses should be conducted to verify the gene controlling cold tolerance at the bud burst stage.
Expression patterns of three candidate genes. a LOC_Os01g55510; b LOC_Os01g55350; c LOC_Os01g55560. Total RNA was isolated from shoot under normal growth conditions and after 1 day cold stress conditions, respectively. Mean and SD values in qRT-PCR analysis were obtained in one experiment with three biological replicates. Variety accessions, S1: CH406; S2: CH314; S3: CH133; S4: CH136; T1: CH191; T2: CH202; T3: CH244; T4: CH307. Different letters indicate significant difference at 1% level (Duncan’s multiple range test)
Discussion
Phenotypic assessment of indica panel
Low temperature stress during the bud burst stage is an important factor affecting rice cultivation in southern China, especially in direct-sowing regions. Cold tolerance is a complicated trait controlled by many genes. To study the genetic basic of cold tolerance in rice, the most important aspect is phenotypic identification. Many of the methods used to assess cold tolerance in rice in previous studies were unsuitable for use in our study, because indica varieties are very sensitive to low-temperature stress. Several studies demonstrated that germinated seeds should be exposed to 5 °C for about 10 days, followed by 7 days of recovery, and subsequently the seed survival rate was estimated. However, this method did not reveal differences among indica varieties because almost all of them died. To reveal differences in cold tolerance among indica varieties, we used a shorter (5-day) low-temperature treatment. After recovery, the survival rate showed a continuous distribution ranging from 0 to 100% in the indica population (Fig. 3c, d). In case the SR data were not useful for GWAS, we also scored SD value and found that it may be more informative than the SR data (Fig. 3a, b). However, comparison of the GWAS results for the SD and SR data showed that they were similar (Fig. 5; Table 1), confirming that SR and SD data were both useful for GWAS.
Unlike the SR value, we scored the cold tolerance of each seed of every variety, with the average score providing the final SD score. Although SD was correlated with SR, the SD was more informative than the SR. Firstly; SD divided cold tolerance into five grades, while SR divided it into only two grades, alive or dead. Therefore, SD provided a more accurate measure of cold tolerance than SR. Secondly, SD provided information about seed purity. If large variations in cold tolerance were detected in one variety, it may indicate that the seeds were not pure and the variety should not be used in the GWAS.
Population structure of indica rice from China
For rice, GWAS is considered as the useful method to reveal complicated genetic variations, but population structure is an important factor that can affect the GWAS results and increase the false positive rate. In our study, the population was composed of 249 indica varieties from China. The genetic structure analysis divided the population into three groups. The result was subsequently supported by the results of the PCA and the NJ tree (Fig. 4). However, all these varieties were in the same subspecies, two major PCs in PCA, PC1 and PC2, accounted for only 9.7 and 5.3% of the genetic variation, respectively, and the three subgroups showed no significant separation in NJ tree and PCA. So there was no strong genetic structure in the indica panel. The relative kinship analysis revealed low relatedness in the panel. Finally, we used the PCA and kinship coefficient to correct the GWAS results.
Comparison between GWAS results in this study and those reported previously
Cold tolerance is a very complicated character in rice. Many QTLs have been identified in rice using the bi-parental mapping strategy. Recently, with the wide use of GWAS to dissect cold tolerance in rice, many relevant loci have been detected. However, indica population panels have rarely been used in GWAS for cold tolerance at the bud burst stage. Moreover, we did not detect cloned genes identified as being related to cold tolerance in previous studies. There are several possible explanations for this. First, all of the other genes related to cold tolerance were not identified at the bud burst stage. Cold tolerance in rice at different stages might be related to different genetic mechanisms. Secondly, rare variation was observed in these regions in indica varieties, many cold-tolerant QTLs were from japonica varieties, it can hardly detected in indica varieties. Although we did not identify specific genes related to cold tolerance in our study, most loci detected overlapped with QTLs reported previously. Among these QTLs, most of them have been detected in bi-parental populations derived from a cross between japonica and indica varieties. Therefore, the alleles of the QTLs that increased cold tolerance were mainly from japonica. The possible reason might be that indica rice was developed from crosses between japonica rice and local wild rice (Huang et al. 2012a); therefore, indica varieties may retain some characters of japonica, such as minor QTLs for cold tolerance.
Identification of candidate gene controlling cold tolerance
Here, we detected a major QTL for cold tolerance on chromosome 1 that was also detected in a bi-parental population in a previous report (Yang et al. 2013). The LD decay analysis indicated that an approximately 579 kb region at the associated locus was a candidate region for further study (Fig. 6). Gene expression pattern analyses are useful to identify candidate gene(s). On the basis of GO enrichment and gene functional annotation results, and gene expression profiles before and after cold treatment in rice (Shen et al. 2014), we selected 18 candidate genes for gene expression analyses. These are common methods to verify the function of genes identified in GWAS, especially those related to abiotic stress. After filtering using these methods, only three genes remained as candidate genes for cold tolerance. Two of the three genes involved in microtubule-based process, and another one associated with glycolytic pathway and expressed in response to abiotic stress. Recently, many novel genes have been cloned as a result of GWAS (Duan et al. 2017, Si et al. 2016; Yano et al. 2016). However, in our study, because the genotype was from the 5K SNPs array, the SNPs density was insufficient to cover every gene. Moreover, different from re-sequencing, we could not compare the sequences of candidate genes between all cold-tolerant and cold-sensitive varieties. Therefore, further analyses are required to identify which gene is related to cold tolerance. The application of GWAS to analyze cold tolerance will be helpful not only for developing molecular markers for use in rice breeding programs, but also for dissecting the genetic basis of cold tolerance in Chinese indica rice.
Author contribution statement
YY AND XW conceived and designed research. YY, MZ and JY conducted experiments. YY, QX, YF and XY performed the phenotypic identification. YY, HY and YW analyzed the data. YY and MZ wrote the manuscript. XW and YW helped to revise the manuscript. All the authors read and approved the final manuscript.
References
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Andaya VC, Mackill DJ (2003) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica x indica cross. Theor Appl Genet 106:1084–1090
Cheng SH et al (2007) Super hybrid rice breeding in China: achievements and prospects. J Integr Plant Biol 49:805–810
Duan P et al (2017) Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant 10:685–694
Fujino K, Matsuda Y (2010) Genome-wide analysis of genes targeted by qLTG3–1 controlling low-temperature germinability in rice. Plant Mol Biol 72:137–152
Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M (2008) Molecular identification of a major quantitative trait locus, qLTG3–1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA 105:12623–12628
Fujino K, Obara M, Shimizu T, Koyanagi KO, Ikegaya T (2015) Genome-wide association mapping focusing on a rice population derived from rice breeding programs in a region. Breed Sci 65:403–410
Han LZ, Zhang SY (2004) Methods of characterization and evaluation of cold tolerance in rice. J Plant Genet Resour 5:75–80
Han LZ, Zhang YY, Qiao YL, Cao GL, Zhang SY, Kim JH, Koh HJ (2006) Genetic and QTL analysis for low-temperature vigor of germination in rice. Acta Genet Sin 33:998–1006
Huang XH et al (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–976
Huang X et al (2012a) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Huang XH et al (2012b) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44:32–39
Huang XH et al (2016) Genomic architecture of heterosis for yield traits in rice. Nature 537:629
Jiang L, Liu SJ, Hou MY, Tang JY, Chen LM, Zhai HQ, Wan JM (2006) Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.). Field Crops Res 98:68–75
Jörgens CI, Grünewald N, Hülskamp M, Uhrig JF (2010) A role for ABIL3 in plant cell morphogenesis. Plant J 62:925–935
Kang HM et al (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42:348–354
Kuroki M, Saito K, Matsuba S, Yokogami N, Shimizu H, Ando I, Sato Y (2007) A quantitative trait locus for cold tolerance at the booting stage on rice chromosome 8. Theor Appl Genet 115:593–600
Lee MH (2001) Low temperature tolerance in rice: the Korean experience. In: Increased lowland rice production in the Mekong Region: proceedings of an international workshop held in Vientiane, Laos, 30 October–2 November 2000
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu FX et al (2013) Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol Plant 6:757–767
Lv Y, Guo ZL, Li XK, Ye HY, Li XH, Xiong LZ (2016) New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis. Plant Cell Environ 39:556–570
Ma Y et al (2015) COLD1 confers chilling tolerance in rice. Cell 160:1209–1221
Nagamine T (1991) Genic control of tolerance to chilling injury at seedling stage in rice, Oryza sativa L. Ikushugaku Zasshi 41:35–40
Pan YH et al (2015) genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. Plos One 10:e0120590
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190
Pradhan SK et al (2016) Population structure, genetic diversity and molecular marker-trait association analysis for high temperature stress tolerance in rice. Plos One 11:e0160027
Saito K, Hayano-Saito Y, Maruyama-Funatsuki W, Sato Y, Kato A (2004) Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor Appl Genet 109:515–522
Saito K, Hayano-Saito Y, Kuroki M, Sato Y (2010) Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci 179:97–102
Sánchez R, Flores A, Cejudo FJ (2006) Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 223:901–909
Schlappi MR et al (2017) Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA mini-core collection. Front Plant Sci 8:957
Shakiba E et al (2017) Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. Plos One 12:e0172133
Shen CX et al (2014) Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. J Plant Biol 57:337–348
Shirasawa S, Endo T, Nakagomi K, Yamaguchi M, Nishio T (2012) Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar, ‘Lijiangxintuanheigu’, in rice, Oryza sativa L. Theor Appl Genet 124:937–946
Si L et al (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet 48:447–456
Sipaseuth et al (2007) Opportunities to increasing dry season rice productivity in low temperature affected areas. Field Crops Res 102:87–97
Sthapit BR, Witcombe JR (1998) Inheritance of tolerance to chilling stress in rice during germination and plumule greening. Crop Sci 38:660–665
Suh JP et al (2010) Identification and analysis of QTLs controlling cold tolerance at the reproductive stage and validation of effective QTLs in cold-tolerant genotypes of rice (Oryza sativa L.). Theor Appl Genet 120:985–995
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Wang D et al (2016) Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice 9:61
Xu LM, Lei Z, Zeng YW, Wang FM, Zhang HL, Shen SQ, Li ZC (2008) Identification and mapping of quantitative trait loci for cold tolerance at the booting stage in a japonica rice near-isogenic line. Plant Sci 174:340–347
Xu Q et al (2016) The genetic diversity and structure of indica rice in China as detected by single nucleotide polymorphism analysis. BMC Genet 17:53–60
Yang ZM, Huang DQ, Tang WQ, Zheng Y, Liang KJ, Cutler AJ, Wu WR (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. Plos One 8:e68433
Yano K et al (2016) Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet 48:927
Zhang Q, Chen Q, Wang S, Hong Y, Wang Z (2014) Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 7:24
Zhao J et al (2017) A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnol J 15:1141–1148
Zhou L et al (2012) Characterization and identification of cold tolerant near-isogenic lines in rice. Breed Sci 62:196–201
Zhu YJ et al (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. Plos One 10:e0145704
Acknowledgements
The present investigation is financially supported by National Natural Science Foundation of China (31600999) and Chinese Academy of Agricultural Sciences (CAAS-ASTIP-201X-CNRRI and CAAS-XTCX2016016).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest declared
Additional information
Communicated by Mark C. Jordan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Comparison of –log10(P) value of 10 common QTLs between SR and SD (TIF 147 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zhang, M., Ye, J., Xu, Q. et al. Genome-wide association study of cold tolerance of Chinese indica rice varieties at the bud burst stage. Plant Cell Rep 37, 529–539 (2018). https://doi.org/10.1007/s00299-017-2247-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2247-4