Abstract
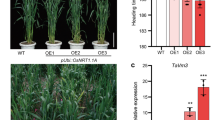
A full-length cDNA clone of OsFAD2, which encodes a Δ-12 fatty acid desaturase, the key enzyme for the conversion of oleic acid (18:1) into linoleic acid (18:2), was isolated from rice (Oryza sativa ssp. japonica) leaves. The deduced amino acid sequence of OsFAD2 displayed three histidine boxes characteristic of all membrane-bound desaturases, and possessed a C-terminal signal for endoplasmic reticulum retention. Phylogenetic analysis showed that OsFAD2 is grouped within plant housekeeping FAD2 sequences. Expression analysis by real-time PCR showed that the gene is expressed in all tissues of rice tested, including root, seed, stem, and leaf. In situ hybridization showed that OsFAD2 mRNA accumulated in leaf mesophyll cells and in root epidermis cells when exposed to 15°C for 4 days in dark conditions. When OsFAD2 was expressed in Saccharomyces cerevisiae, the cells could convert oleic acid to linoleic acid, which wild-type yeast cells cannot do, suggesting that the isolated gene encoded a functional FAD2 enzyme. Heterologous expression of OsFAD2 enhanced the yeast cells’ cold tolerance capacity compared to wild-type yeast. OsFAD2 was also shown to be a highly active desaturase when expressed in Xenopus oocytes. In addition, when the OsFAD2 gene was transferred into an Arabidopsis thaliana fad2-1 mutant, it effectively restored wild-type fatty acid composition and growth characteristics. Stress tolerance and light regulatory elements were identified in the predicted promoter of the OsFAD2 gene. Exogenously supplied hormone affected the level of FAD2 mRNA accumulation, accompanied by a change of content of di-unsaturated fatty acid species in rice leaves. Furthermore, OsFAD2 enhanced tolerance to low temperature when overexpressed in rice at the vegetative stage. More importantly, the 35S::OsFAD2 plants showed significantly enhanced cold tolerance at the reproductive stage, increasing grain yield by 46% over controls in the greenhouse under cold conditions. These results indicated that OsFAD2 is involved in fatty acid desaturation and maintenance of the membrane lipids balance in cells, and could improve the tolerance of yeast and rice to low temperature stress.



Similar content being viewed by others
References
Adams A, Gottschling DE et al (1998) Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Amiard V, Demmig-Adams B et al (2007) Role of light and jasmonic acid signaling in regulating foliar phloem cell wall ingrowth development. New Phytol 173(4):722–731
Baker SS, Wilhelm KS et al (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24(5):701–713
Browse J, Xin Z (2001) Temperature sensing and cold acclimation. Curr Opin Plant Biol 4(3):241–246
Changhua J, Jianyao XU et al (2009) A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell Environ 32(8):1046–1059
Cleaver OB, Patterson KD et al (1996) Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development 122(11):3549–3556
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Covello PS, Reed DW (1996) Functional expression of the extraplastidial Arabidopsis thaliana oleate desaturase gene (FAD2) in Saccharomyces cerevisiae. Plant Physiol 111(1):223–226
Dominguez T, Hernandez ML et al (2010) Increasing omega-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol 153(2):655–665
Dyer JM, Mullen RT (2001) Immunocytological localization of two plant fatty acid desaturases in the endoplasmic reticulum. FEBS Lett 494(1–2):44–47
Heppard EP, Kinney AJ et al (1996) Developmental and growth temperature regulation of two different microsomal omega-6 desaturase genes in soybeans. Plant Physiol 110(1):311–319
Hernandez ML, Mancha M et al (2005) Molecular cloning and characterization of genes encoding two microsomal oleate desaturases (FAD2) from olive. Phytochemistry 66(12):1417–1426
Hiei Y, Ohta S et al (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6(2):271–282
Kajiwara S, Shirai A et al (1996) Polyunsaturated fatty acid biosynthesis in Saccharomyces cerevisiae: expression of ethanol tolerance and the FAD2 gene from Arabidopsis thaliana. Appl Environ Microbiol 62(12):4309–4313
Kamiya T, Tanaka M, Mitani N, Ma JF, Maeshima M, Fujiwara T (2009) NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J Biol Chem 284:2114–2120
Kargiotidou A, Deli D et al (2008) Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J Exp Bot 59(8):2043–2056
Khodakovskaya M, Li Y et al (2005) Effects of cor15a-IPT gene expression on leaf senescence in transgenic Petunia × hybrida and Dendranthema × grandiflorum. J Exp Bot 56(414):1165–1175
Khodakovskaya M, McAvoy R et al (2006) Enhanced cold tolerance in transgenic tobacco expressing a chloroplast omega-3 fatty acid desaturase gene under the control of a cold-inducible promoter. Planta 223(5):1090–1100
Knutzon DS, Thompson GA et al (1992) Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proc Natl Acad Sci USA 89(7):2624–2628
Liu KH, Tsay YF (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22(5):1005–1013
Lu C, Wallis JG et al (2007) An analysis of expressed sequence tags of developing castor endosperm using a full-length cDNA library. BMC Plant Biol 7:42
Ma JF, Yamaji N, Mitani N, Xu X-Y, Su Y-H, McGrath SP, Zhao F-J (2008) Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA 105(29):9931–9935
Martínez-Rivas JM, Sperling P et al (2001) Spatial and temporal regulation of three different microsomal oleate desaturase genes (FAD2) from normal-type and high-oleic varieties of sunflower (Helianthus annuus L.). Mol Breed 8(2):159–168
McCartney AW, Dyer JM et al (2004) Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J 37(2):156–173
Mietkiewska E, Brost JM et al (2006) A Tropaeolum majus FAD2 cDNA complements the fad2 mutation in transgenic Arabidopsis plants. Plant Sci 171(2):187–193
Mikkilineni V, Rocheford TR (2003) Sequence variation and genomic organization of fatty acid desaturase-2 (fad2) and fatty acid desaturase-6 (fad6) cDNAs in maize. Theor Appl Genet 106(7):1326–1332
Miquel M, Browse J (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl- phosphatidylcholine desaturase. J Biol Chem 267(3):1502–1509
Niu B, Ye H et al (2007) Cloning and characterization of a novel Δ12-fatty acid desaturase gene from the tree Sapium sebiferum. Biotechnol Lett 29(6):959–964
Oh S-J, Kim YS et al (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150(3):1368–1379
Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7(7):957–970
Okuley J, Lightner J et al (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6(1):147–158
Qiu X, Reed DW et al (2001) Identification and analysis of a gene from Calendula officinalis encoding a fatty acid conjugase. Plant Physiol 125(2):847–855
Rodriguez-Vargas S, Sanchez-Garcia A et al (2007) Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol 73(1):110–116
Shanklin J, Cahoon EB (1998) Desaturation and related modifications of fatty acids 1. Annu Rev Plant Physiol Plant Mol Biol 49(1):611
Teixeira MC, Coelho N et al (2009) Molecular cloning and expression analysis of three omega-6 desaturase genes from purslane (Portulaca oleracea L.). Biotechnol Lett 31(7):1089–1101
Tong Y, Zhou J-J, Li Z, Miller AJ (2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41(3):442–450
Upchurch R (2008) Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol Lett 30(6):967–977
Wang J, Ming F et al (2006) Characterization of a rice (Oryza sativa L.) gene encoding a temperature-dependent chloroplast [omega]-3 fatty acid desaturase. Biochem Biophys Res Commun 340(4):1209–1216
Watanabe K, Oura T et al (2004) Yeast Δ12 fatty acid desaturase: gene cloning, expression, and function. Biosci Biotechnol Biochem 68(3):721–727
Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7(5):217–224
Wenbin Z, Yuxiang C et al (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58(1):82–96
Wu J, Lightner J et al (1997) Low-temperature damage and subsequent recovery of fab1 mutant Arabidopsis exposed to 2[deg]C. Plant Physiol 113(2):347–356
Zhang D, Pirtle IL et al (2009) Identification and expression of a new delta-12 fatty acid desaturase (FAD2–4) gene in upland cotton and its functional expression in yeast and Arabidopsis thaliana plants. Plant Physiol Biochem 47(6):462–471
Acknowledgments
The study was supported by the National High Technology Research and Development Program of China (2008AA10Z116) and the Ministry of Agriculture of China (2008ZX08009-001 and 2009ZX08009-061B) for Feng Ming.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11032_2011_9587_MOESM1_ESM.docx
Supplemental Fig. 1 Alignment of predicted amino acid sequences of plant FAD2 polypeptides using ClustalW program. Identical amino acid residues are indicated by reverse contrast. The deduced amino acid sequences compared are from: Triadica sebifera (TsFAD2, DQ903666); Glycine max (GmFAD2, DQ532371); Arabidopsis thaliana (AtFAD2, AAA32782), Calendula officinalis (CoFAD2, AF343065), Punica granatum (PgFAD2, AY178447), Zea mays (ZmFAD2, EF687907), Sorghum bicolor (SbFAD2; EF206347), and Oryza sativa (OsFAD2, FJ768953) FAD2 homologs. Boxes represent histidine motifs. The cDNA sequence corresponding to OsFAD2 has been deposited in the GenBank database with the accession no. FJ768953 (DOCX 130 kb)
11032_2011_9587_MOESM2_ESM.docx
Supplemental Fig. 2 Phylogenetic analysis of plant FAD2 enzymes. The dendrogram was arbitrarily rooted with the Arabidopsis thaliana FAD3 sequence. Distances along the horizontal axes are proportional to sequence differences. Position of the rice microsomal oleate desaturase gene is triangular. Accession numbers of the different desaturases included in the analysis: Arabidopsis thaliana (AtFAD2, L26296), Gossypium hirsutum (GhFAD2-2, Y10112) Triadica sebifera (TsFAD2, DQ903666), Brassica carinata (BcFAD2, AF124360), Cucurbita pepo (CpeFAD2, AY525163), Brassica juncea (BjFAD2, X91139), Brassica campestris (BrFAD2, AJ459107), Brassica napus (BnFAD2, AF243045), Punica granatum (PgFAD2, AY178447), Glycine max (GmFAD2-2, L43921), Glycine max (GmFAD2-3, DQ532371), Gossypium hirsutum (GhFAD2-3, AF331163), Vernonia galamensis (VgFAD2-2, AF188264; VgFAD2-1, AF188263), Crepis palaestina (CpaFAD2, Y16284), Calendula officinalis (CoFAD2, AF343065), Borago officinalis (BoFAD2, AF074324), Olea europaea (OepFAD2, AY733077), Spinacia oleracea (SoFAD2, AB094415), Petroselinum crispum (PcFAD2,U86072), Helianthus annuus (HaFAD2-2, AF251843; HaFAD2-3, AF251844), Persea americana (PamFAD2, AY057406), Glycine max (GmFAD2-1A, L43920; GmFAD2-1B, AB188251), Arachishypogaea (AhFAD2B, AF272950; AhFAD2A, AF030319), Arachis ipaensis (AiFAD2, AF272952), Arachis duranensis (AdFAD2, AF272951), Olea europaea (OepFAD2-1, AY733076), Sesamum indicum (SiFAD2, AF192486), Gossypium hirsutum (GhFAD2-1, X97016), Solanum commersonii (ScFAD2, X92847), Vernicia fordii (VfFAD2, AF525535), Arabidopsis thaliana (AtFAD3, NM_128552) (DOCX 69 kb)
Rights and permissions
About this article
Cite this article
Shi, J., Cao, Y., Fan, X. et al. A rice microsomal delta-12 fatty acid desaturase can enhance resistance to cold stress in yeast and Oryza sativa . Mol Breeding 29, 743–757 (2012). https://doi.org/10.1007/s11032-011-9587-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-011-9587-5