Abstract
In the past three decades, biomedical engineering has emerged as a significant and rapidly growing field across various disciplines. From an engineering perspective, biomaterials, biomechanics, and biofabrication play pivotal roles in interacting with targeted living biological systems for diverse therapeutic purposes. In this context, in silico modelling stands out as an effective and efficient alternative for investigating complex interactive responses in vivo. This paper offers a comprehensive review of the swiftly expanding field of machine learning (ML) techniques, empowering biomedical engineering to develop cutting-edge treatments for addressing healthcare challenges. The review categorically outlines different types of ML algorithms. It proceeds by first assessing their applications in biomaterials, covering such aspects as data mining/processing, digital twins, and data-driven design. Subsequently, ML approaches are scrutinised for the studies on mono-/multi-scale biomechanics and mechanobiology. Finally, the review extends to ML techniques in bioprinting and biomanufacturing, encompassing design optimisation and in situ monitoring. Furthermore, the paper presents typical ML-based applications in implantable devices, including tissue scaffolds, orthopaedic implants, and arterial stents. Finally, the challenges and perspectives are illuminated, providing insights for academia, industry, and biomedical professionals to further develop and apply ML strategies in future studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In recent decades, cutting-edge engineering disciplines have increasingly converged with biological systems, significantly impacting the healthcare industry. Biomedical engineering, in particular, has rapidly emerged as an innovative and multidisciplinary field that has drawn extensive interest from research communities. Figure 1a illustrates some fast-growing disciplines within biomedical engineering, including biomaterials [1,2,3], biomechanics/mechanobiology [4,5,6,7], and biofabrication [8, 9]. These fields seamlessly integrate material sciences, mechanics, mathematics, physics, chemistry, computer science, and advanced manufacturing with biological sciences and clinical medicine, resulting in substantial benefits to human healthcare and socio-economic systems.
Outline of review contents. a Machine learning (ML) in biomaterials, biomechanics/mechanobiology, and biofabrication. Created with BioRender.com. b The number of publications related to ML or data-driven approaches in conventional engineering since 2010. c The number of publications related to ML or data-driven approaches in biomedical engineering fields since 2010
Biomaterials present a broad class of natural and synthetic materials that are able to intimately interact with living biological systems. They exhibit multifunctionalities crucial for immune system support, cell interactions, and response to chemical, physical, and mechanical conditions within a local biological environment [10, 11]. So far, polymers, metals, ceramics, and composites are amongst the most typical biomaterials extensively utilised in various biomedical applications, including implantable and wearable devices [12, 13], regenerative medicine [14] and drug delivery [15]. Figure 2a depicts biomaterials employed in regenerative medicine for cardiac, bone, and skin tissue engineering, as well as drug delivery systems. These materials encompass a variety, incorporating polymeric, inorganic, and lipid-based nanoparticles. For decades, substantial efforts have been dedicated to exploration of novel biomaterials with desired chemical, physical, mechanical, and biological properties [1, 2]. Conventional routes for discovering and developing new biomaterials often relied heavily on trial-and-error experimental tests, proving workable but costly and time-consuming. In order to expedite the experimental processes and enhance success rates, advanced computational techniques have been widely employed as complementary approaches in modern biomaterial design [16, 17], which have proven effective in disclosing how material constituents and structures influence multifunctional properties from macroscale to nanoscale, thus offering compelling opportunities to achieve desired material performances as well as customised functionalities for patient-specific applications.
Biomechanics and mechanobiology often play critical roles in comprehending the responses and adaptation of living tissues to local environmental changes induced by prosthetic and therapeutic treatments. These disciplines closely engage with forces, deformation, stiffness, permeability, and other physical fields such as temperature and electromagnetic signals, exerting significant control over load-bearing characteristics and biotransportation across molecular, cellular, tissue, and organ levels [18,19,20,21,22,23,24], as depicted in Fig. 2b. While mechanobiology is closely correlated with biomechanics, it places a greater emphasis on actively regulating in vivo tissues or cellular behaviours such as tissue growth/remodelling and cell differentiation/proliferation [25,26,27,28,29]. As shown in Fig. 2b, mechanobiology regulates cellular behaviours with mechanical stimuli, thereby affecting osteoclasts, monocytes, pre-osteoblasts, osteoblasts, and osteocytes processes for bone adaptation and remodelling. The development of biomechanical and mechanobiological modelling techniques has been rapid, facilitated by advances in computational and data sciences integrated with innovative non-invasive imaging technologies [18, 30,31,32]. In both preclinical research and clinical trials, biomechanical and mechanobiological modelling have made significant contributions and theoretical breakthroughs in unveiling the complex relationships among a range of factors in engineering, biomaterials, and biomedicine. For example, disciplines like tissue engineering [33, 34], orthopaedics [35, 36], oromaxillofacial reconstruction [37,38,39], cardiovascular and pulmonary systems [40, 41] have widely embraced biomechanical and mechanobiological modelling techniques, thereby advancing multidisciplinary knowledge in this rapidly emerging field.
Applications of biomaterials, biomechanics/mechanobiology, and biofabrication. a Biomaterials for implantable and wearable devices, including prosthetics, dental implants, scaffolds, and stents. Created with BioRender.com. b Illustration of biomechanics/mechanobiology for bone tissue engineering. Created with Biorender.com. c Biofabrication using bioink. Reproduced with permission. Copyright 2016, Elsevier [42]
Biofabrication encompasses a versatile set of advanced manufacturing techniques for creating non-living biomaterials, living constructs (cells or tissues), and hybrid components [43]. Recently, additive manufacturing (AM) has been rapidly developed to craft novel and sophisticated implantable or wearable devices, including bone scaffolds [44], fixation plates [45], dental implants [46], and smart surfaces [47]. For non-living materials, biomanufacturing can be mainly classified by different construction processes, such as fused deposition modelling (FDM), stereolithography (SLA) for polymers [48], selective laser sintering (SLS), and electron beam melting (EBM) for metals and alloys [49], as well as vat photopolymerisation and binder jetting (BJ) for bioceramics [50]. A more recent and notable development in this field is bioprinting, accomplished through ink-jet and valve-jet printing techniques that utilise customised bioink, combining living cells/tissues with supporting base materials to directly mimic structures of native organs/tissues [43]. Figure 2c illustrates bioprinting, depicting the creation of a microfibrous scaffold using a composite bioink encapsulating endothelial cells.
Lured by remarkable advances in computational sciences and computer technologies over the past decades, there has been a remarkable increase in data generation. For this reason, how to manage and make use of the unprecedented amount of data has inspired researchers to develop a range of data-driven approaches. Machine learning (ML), as a prominent class of data-driven approaches, employs computational algorithms learned from data, as opposed to experience or theory, to enhance performance in solving specific tasks. It stands as one of the most prevalent computational strategies utilised in a broad range of fields, including image and voice recognition, autonomous driving, online fraud detection, automatic language translation and medical diagnosis [51].
Engineering has undeniably witnessed profound impacts from various ML approaches, with significant efforts dedicated to material sciences [52], computational modelling techniques [33], and AM [53]. Figure 1b depicts the publication trend in these fields from the Web of Science Core Collection since 2010. Each subcategory, namely “machine learning” or “data-driven” combined with “materials”, “mechanics”, and “additive manufacturing/3D printing”, was counted. The publications in these fields have experienced remarkable growth since 2016. Material sciences, in particular, have emerged as the most dynamic discipline, with over 6,500 publications in 2023 alone, and the upward trend is expected to continue in the following years. While the percentages of publications in AM and mechanics are slightly lower than that of materials sciences, they are notably increasing, demonstrating potent applications across diverse disciplines within broad engineering fields.
Nevertheless, despite the unprecedented success of ML in other traditional engineering disciplines, biomedical engineering remains relatively underexplored. Figure 1c analyses the number of publications applying ML in biomaterials, biomechanics/mechanobiology, and biofabrication since 2010. The publications with topics containing keywords such as machine learning/data-driven plus biomaterial, biomechanics, mechanobiology, bioprinting, and biofabrication were counted from the Web of Science Core Collection. A rapid increase in publications has been more evident since 2017, with over 200 articles in 2023 alone. Biomechanics and mechanobiology constitute the largest portion of these publications (60.89%). While the total number of publications in these areas lags behind those shown in Fig. 1b, it reveals significant potential and tremendous opportunities for biomedical engineering to establish a new paradigm.
Therefore, the purpose of this review is to conduct a state-of-the-art evaluation of the development of ML in biomedical engineering and provide insights into its potential applications in relevant areas. We focus on various ML techniques applied in biomaterials, biomechanics/mechanobiology, and biofabrication from an engineering perspective, as outlined in Fig. 1a. It is important to note that these areas are highly interdisciplinary in nature; and other important disciplines, such as biophysics, biochemistry, and biology, may not have been reviewed comprehensively here.
The remaining sections are organised as follows. Section 2 briefs the ML approaches commonly used in the reviewed studies. Sections 3–5 review ML in disciplines of biomaterials, biomechanics/mechanobiology, and biofabrication, respectively. Section 6 focuses on typical applications in bone scaffolds, orthopaedic/dental implants, and arterial stents. Section 7 discusses the challenges and perspectives, followed by a conclusion in Sect. 8.
2 Machine Learning Approaches
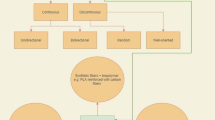
Machine learning integrates principles from statistics, neural networks, optimisation theory, computer science, system identification, and various other fields. Its overarching goal is to simulate or implement human learning behaviours, enabling the continuous improvement and reorganisation of known skills [54]. According to the differences in learning manners, ML can be categorised into supervised learning [55], unsupervised learning [56], semi-supervised learning [57], and reinforcement learning [58], as depicted in Fig. 3. Given the dynamic nature of ML techniques, the algorithms mentioned here may not represent an exhaustive enumeration of ML methods. Nevertheless, this section briefly reviews some typical ML models extensively utilised in biomedical engineering in open literature.
2.1 Supervised Learning
Supervised learning utilises labelled training data to establish a mapping with new instances. Labels for training samples must be provided, and higher labelling accuracy generally leads to more effective learning outcomes. Supervised learning models aim to find an implicit functional relationship between input and output data based on given knowledge, making it suitable for classification and regression problems. Commonly used supervised learning models are summarised below.
2.1.1 K-Nearest Neighbour (KNN)
KNN, proposed by Fix et al. [59] and enhanced by Cover and Hart [60], stands as a prominent algorithm for classification and pattern regression [61]. It operates on the premise that similar instances are proximate, allowing the identification of new input features by calculating distances to existing sample data. Subsequently, inputs are classified into the nearest category. The essence of KNN lies in measuring distances between tested and training samples. In this context, Surya et al. [62] conducted a comprehensive review of KNN performance using various distance measures. The advantages of KNN stem from its simplicity in developing a training model [63]. Furthermore, the need for parametric regulation of complex models is eliminated [64]. However, it is worth noting that KNN may exhibit reduced efficiency with a substantial volume of sampling data. Nevertheless, KNN has found widespread applications in recommendation systems, text mining, finance, agriculture [65], and medical-related prediction [66,67,68], among other domains.
2.1.2 Decision Tree (DT)
DT is a fundamental method for classification and regression, encompassing both a classification tree and a regression tree. Among the most classical algorithms are ID3, C4.5, and C5.0 [69,70,71]. DT manifests a tree structure where internal nodes represent attribute tests, branches depict the outputs of these tests, and leaf nodes house the classification labels. It can be perceived as a compilation of if–then rules or as a conditional probability distribution defined in feature and classification spaces. Learning steps for DT typically involve feature selection, generation of DT structures, and model trimming. Due to its visual structure, DT is easily comprehensible and widely applicable across various domains, including data analysis in biomedical fields [72,73,74,75,76,77,78].
2.1.3 Random Forest (RF)
RF, proposed by Breiman [79], is composed of multiple DTs constructed with random feature subsets. Therefore, RF is recognised as an ensemble algorithm. Each DT functions as a classifier, and RF integrates the classification results from all DTs. The classification with the highest proportion is determined as the final output result. Due to the randomness inherent in DT construction, the common issue of overfitting can be alleviated to a considerable extent [80]. The RF algorithm has found certain applications in various biomedical fields to date [81,82,83,84].
2.1.4 Naïve Bayesian (NB)
Based upon the Bayes theorem, the NB algorithm classifies data samples using knowledge of probability statistics [85]. Unlike Decision Trees (DT), NB is firmly rooted in a more robust mathematical foundation. It assumes that all attributes are independent, allowing the NB algorithm to learn the joint probability distribution from input samples to output data. After training the NB model, it can generate output results with the greatest posterior probability when given input values. However, meeting the requirement of independence for dataset attributes can be challenging in many cases. Consequently, numerous studies have been conducted to address this assumption by considering attribute weighting, attribute selection, and structure extension [86]. The NB algorithm has extensive applications in various biomedical areas [87, 88].
2.1.5 Support Vector Machine (SVM)
SVM [89] has found extensive applications in statistical classification and regression analysis. In general, it belongs to a linearised classifier aiming to maximise the interval in feature spaces by solving convex quadratic programming problems. SVM maps the vectors of samples to a higher-dimensional space, where a hyperplane best separates two groups of mapped vectors. The advantages of SVM lie in its ability to learn data samples with good reproducibility and accuracy, ensuring that the model is generic and capable to new data, thereby maximising the proportion of correct labels [90]. SVM has been applied across various fields, such as text classification [91], image classification [92], biological sequence analysis, biological data mining [93], biomechanics [94,95,96,97,98], regenerative medicine [99, 100], and more.
2.1.6 Logistic Regression and Linear Regression
Logistic regression and linear regression [101, 102] are similar in many aspects, both falling under the umbrella of generalised linear models. The primary distinction lies in the types of their outputs. If the output is continuous, it is referred to as multiple linear regression and is employed to address regression problems. On the other hand, if the output follows a binomial distribution, it is recognised as logistic regression and is frequently used for classification issues. The process for both algorithms involves selecting data sets and output variables of interest, specifying a ML model, training the model parameters, and conducting model evaluation and validation [103]. These methods are fairly popular and widely utilised in various fields, such as biometrics [104], finance prediction [105], disease diagnosis [106], etc.
2.1.7 Backpropagation Neural Network (BPNN)
An artificial neural network (ANN) [107] is a complex system comprising numerous nodes (neurons) interconnected by pathways designed to emulate human brain functions. The outputs of an ANN are determined by various factors, including network structure, connection methods, weights, and activation functions. The weights in an ANN require training based on a sufficient dataset. Typically, an ANN consists of one input layer, one or more hidden layers, and one output layer. Among the learning frameworks for ANNs, it appears that the backpropagation neural network (BPNN) stands out. In a BPNN, initial weights are assigned randomly, and output data are generated as input data traverse the entire ANN model. These output data are then compared with known correct results, and any discrepancies are fed back from the output layer to the input layer to iteratively update the neural weights of the ANN model. Numerous techniques have been developed to optimise these weights, including the Levenberg–Marquardt (LM) method [108], scaled conjugate gradient (SCG) [109], one-step secant (OSS) [110], and others. This iterative process continues until the output errors fall within an acceptable tolerance.
2.1.8 Convolutional Neural Networks (CNN)
Convolutional neural network (CNN) [111] is one of the representative algorithms for deep learning, crafted to mimic the visual perception processes observed in animals. It is adaptable to both supervised and unsupervised learning paradigms. In this architecture, key components include input and hidden layers, with the hidden layers typically comprising convolutional layers, pooling layers, and fully connected layers. Convolutional layers play a pivotal role in extracting features from input data samples, while pooling layers serve to filter these features and convey new information to the fully connected layer. Regardless of implementing the supervised or unsupervised learning frameworks, CNNs leverage the concept of transferring error information backwards, akin to BPNN, facilitating the iterative adjustment of model parameters to enhance learning.
2.2 Unsupervised Learning
In contrast to supervised learning, unsupervised learning models are designed to unveil inherent structures or patterns within unlabelled data samples [112]. Despite this capability, the absence of corrective mechanisms inherent in supervised approaches makes it challenging to ensure the reasonability of learning models during the learning process. Unsupervised learning excels in discerning underlying laws among input data samples. Once trained and verified, these models find proper application in novel scenarios. Two prevalent techniques employed in unsupervised learning are clustering and dimension reduction [113]. Notable algorithms within this domain include K-means [114], Self-Organising Map (SOM) [115], Principal Component Analysis (PCA) [116], and CNN [117] as follows.
2.2.1 K-means
The K-means algorithm [118] is one of the most popular unsupervised ML models for its simplicity. The fundamental concept behind K-means is to categorise samples into the most similar groups based on the distances between each sample and category centre. Upon introduction of new samples into each cluster, the category centres undergo updates. The final classification of each sample is determined through iterative processes, ceasing when no further changes in category centres occur. However, due to the necessity of calculating distances between samples and all category centres, the K-means algorithm may exhibit sluggish performance when applied to large-scale datasets. Additionally, its sensitivity to noise may cause category centres to deviate to some extent from the correct ones in certain cases [114].
2.2.2 Self-Organising Map (SOM)
SOM [119] can be conceptualised as a straightforward neural network consisting only of an input layer and an output layer without hidden layers. SOM endeavours to map a dataset from any dimension into a one-dimensional or two-dimensional space by adaptively performing a transformation in an organised manner. Employing competitive learning, the winning neuron which is most closely aligned with the input data, is activated, prompting updates to the parameters of nodes neighbouring the activated neuron in terms of their distances from the winner. SOM facilitates the visualisation of database structures in a single image and has found extensive applications in clustering and data mining across various domains, including finance, industry, biomedical science, and more [120,121,122,123,124].
2.2.3 Principal Component Analysis (PCA)
PCA [125] is one of the most frequently utilised linear dimension reduction algorithms. It achieves the transformation of high-dimensional datasets into a lower-dimensional space through various forms of linear projection. PCA aims to maximise the variance of datasets in the projected dimensional space, enabling the utilisation of fewer dimensions while preserving the essential characteristics of the original datasets [126]. As a linear dimensional reduction method, PCA minimises the loss of features from the original dataset. Essentially, PCA seeks to distil key information from data samples, facilitating a simplified characterisation of the datasets [127]. Furthermore, PCA is adept at noise reduction within datasets and contributes on computational efficiency. This method finds applications in various biomedical areas [95, 128, 129].
2.2.4 Convolutional Neural Network (CNN) for Unsupervised Learning
In scenarios with limited labelled samples, CNN can be extended to the realm of unsupervised learning. Several models have been proposed in this context, including Variational Autoencoders (VAE) [130], Convolutional Restricted Boltzmann Machines [131], Deep Convolutional Generative Adversarial Networks [132], and so on.
2.3 Semi-supervised Learning
Semi-supervised learning [133] emerges as a valuable approach in scenarios featuring both labelled and unlabelled data samples, amalgamating principles from supervised and unsupervised learning. Often, full supervision proves unnecessary, and semi-supervised learning offers a less time-consuming and labour-intensive alternative for manually labelling training samples [134, 135]. Generally speaking, this methodology leverages a smaller set of labelled samples alongside a larger pool of unlabelled ones. The inclusion of unlabelled samples mitigates the challenges associated with performance degradation in conventional supervised learning when training samples are insufficient. Prominent algorithms in semi-supervised learning include self-training, semi-supervised support vector machine (S3VM), and graph-based methods.
2.3.1 Self-Training
Self-training represents the simplest method in semi-supervised learning, seeking to augment labelled datasets using unlabelled data samples [136, 137]. The process involves training with a limited number of labelled samples and subsequently labelling unlabelled samples with a well-trained ML model [138]. Given the potential inaccuracy of predictions from a model trained on an insufficient dataset, filtering techniques may be necessary in this context. A key drawback of self-training is the potential introduction of noisy labels by a well-trained ML model, prompting further studies to address this issue [139, 140].
2.3.2 Semi-supervised Support Vector Machine (S3VM)
As proposed by Bennett [57], Semi-Supervised Support Vector Machine (S3VM) extends the conventional SVM method in the realm of semi-supervised learning. In scenarios without unlabelled samples [141], it resembles SVM, aiming to identify a hyperplane with the maximum interval distance between support vectors. When considering unlabelled samples, S3VM attempts to establish a hyperplane that not only separates different types of labelled samples but also navigates through low-density areas in the dataset [142]. In this regard, Ding et al. [143] provide a comprehensive review of mainstream models in semi-supervised support vector machines, including Transductive SVM (TSVM), Laplacian SVM (LapSVM), MeanS3VM, and S3VM based upon cluster kernels.
2.3.3 Graph-Based Methods
Graph-based methods include minute [144], spectral graph transducer [145], Gaussian fields, harmonic function [146], etc. These methods share some similarities with the nearest-neighbour learning algorithms in supervised frameworks, differing in their incorporation of unlabelled samples to enhance ML model performance. Labelled and unlabelled data samples are treated as vertices connected by edge weights. The generation of graph edges, computation of edge weights, and execution of graph-based algorithms constitute crucial steps, with the effectiveness of this semi-supervised ML algorithm reliant on well-defined graph edges and edge weights [147].
2.4 Reinforcement Learning
Reinforcement learning [148] stands as the fourth fundamental category of ML methods, alongside supervised learning, unsupervised learning, and semi-supervised learning. In contrast to supervised learning, which aims to train ML models for producing correct outcomes, reinforcement learning places a strong emphasis on evaluating outcomes through reinforcing signals. Reinforcement learning models evolve through existing experiences and learning from mistakes to achieve improved results. Common reinforcement learning algorithms include Monte Carlo learning and Q-learning.
2.4.1 Monte-Carlo Learning
Monte Carlo learning involves the use of a substantial number of random samples to explore the entire knowledge space by directly learning from the environment [149]. This approach allows the construction of a relatively abstract model using known data samples, with the model parameters determined through the Monte Carlo technique to minimise residuals from original data. The Monte-Carlo algorithm learns from experiences, encompassing the state of samples, actions, and rewards. Upon extracting experiences from samples, reinforcement learning tasks can be addressed based on average sample returns [150, 151]. This type of reinforcement learning algorithm is less sensitive to initial values. However, the convergence of Monte-Carlo learning can be a key issue, and many studies have been carried out to deal with it [152, 153]. This reinforcement learning algorithm exhibits lower sensitivity to initial values. However, achieving convergence in Monte-Carlo learning can be a pivotal challenge, prompting numerous studies in the literature over time [154].
2.4.2 Q-learning
Q-learning, introduced by Watkins and Dayan [155], employs a Q-table as a reference to explore external states and receive rewards until a target state is attained. The training process of Q-learning primarily involves strengthening the ‘brain’ conventionally represented as the ‘Q’ table. It excels in identifying the most efficient path to reach the desired state effortlessly [156]. In comparison to the Monte Carlo reinforcement learning algorithm, Q-learning is more efficient but exhibits higher sensitivity to initial values [157, 158].
3 Machine Learning in Biomaterials
3.1 Overview
Biomaterials encompass versatile subclasses of organic/inorganic biocompatible materials that can intimately interact with living tissues. Apart from their mechanical and physical properties, tissue/cellular interactions with biomaterials have proven critically important [159, 160]. Table 1 summarises some typical biomaterials to illustrate the application spectrum. For example, polymers signify one of the major subclasses, varying from natural biopolymers such as proteins and polynucleotides [161] to synthetic degradable and non-degradable ones [162]. Their applications embrace a fairly broad range, such as connective hard and soft tissues [163], bioinks [43], drug delivery media, and tissue scaffolds [164]. Metals and their alloys are other important subclasses of biomaterials owing to their excellent mechanical properties and inertness [165], which have been well developed as a good alternative for either temporary or permanent replacement of failure tissues. Typical metallic biomaterials include stainless steel, titanium alloys, magnesium alloys, and shape memory alloys, which are widely used in orthopaedic devices [38, 39], dental devices [166,167,168,169,170,171,172,173], wearable devices [13] and arterial stents [41]. Similar to metallic biomaterials, ceramics have been used to replace or restore some diseased hard tissues (e.g., bone and teeth) owing to their excellent mechanical properties, chemical resistance, and transparency [33, 34, 174]. Composite biomaterials, generally composed of two or more materials with different compositions and microstructures, have also been studied and tested in medical applications such as orthopaedic implants and tissue scaffolds [175, 176], which could offer more freedom for engineers to customise various functionalities with ease.
Conventional routes for unravelling novel biomaterials rely on a large number of trial-and-error experiments in vitro and/or in vivo, which are generally time-consuming and uneconomic. Therefore, a so-called “rational design” using computational techniques has been more and more favoured for exploring novel biomaterials recently [212]. Latest advances in ML approaches have inspired biomaterials engineers and developers in rational design for their superior ability to handle a large volume of data over human experience, which has extensively influenced the design philosophy and development of new biomaterials as well as their clinical applications [213,214,215].
The ML applications in biomaterials can be categorised into three typical areas, namely data mining/processing, digital twins, and data-driven design. Data mining/processing allow identifying decisive factors affecting the target biomaterial properties, thus offering an intuitive way for characterising and understanding biomaterials. Digital twins establish quantitative relationships between those determinative factors and desired biomaterial properties, which can play a critical role in obtaining real-time responses of desired performances. Benefiting from the data mining/processing and digital twins, biomaterial design allows better customising those determinative factors for achieving desired and/or optimal functionalities [216, 217]. These three categories play important roles in exploring novel biomaterials, which are analysed in the following subsections. In this section, the analysis was conducted under the thematic umbrellas of biomaterials with data mining/data processing, digital twins, as well as data-driven design as follows.
3.2 Data Mining/Processing for Biomaterials
Biomaterials are generally associated with substantial data acquired from either experimental (in vitro and/or in vivo) tests or in silico modelling. ML techniques such as clustering, classification, and dimensionality reduction can be used to excavate such big data to determine the most relevant and dominant factors for the targeted biomaterial properties. In this aspect, Madiona et al. [177] employed the self-organising map (SOM) method to reduce the dimensionality of data describing surface interactions between polymers and living tissues (Fig. 4a), which provides an effective way to understand the molecular properties of polymer surfaces. In their study, the SOM was constructed with a network size of 8 × 8 and trained through a specified number of iterations, totalling 10,000 epochs. Figure 4a visually represents the data labels with distinct colours: polyethene terephthalate-red, poly(methyl methacrylate)-green, low-density polyethene-blue, poly(caprolactam)-sky blue, poly(undecanoamide)-lavender, poly(lauryllactam)-light yellow, poly(trimethyl-hexamethylene terephthalamide)-dark green, poly(hexamethylene adipamide)-indigo, Poly(hexamethylene azelamide)-dark red, and Poly(hexamethylene dodecanediamide)-light blue. Baier et al. [199] used the K-means clustering method to analyse micropores that could distinctively influence cellular physiology and new bone ingrowth in CaP bioceramics, in which five geometrical parameters (Fig. 4b) in each clustering group were investigated. In Fig. 4b, the three micropore clusters are based on Feret’s diameter and circularity as highlighted in black (1), red (2), and green (3). Shen et al. [189] studied the cell proliferation with titanium dioxide nanotube (TNT) using a DT model, where it was found that cell density and sterilisation could simultaneously impact the cell proliferation on various TNTs (Fig. 4c). The study involved a comparison between predicted and measured cell proliferation values using Decision Trees (DT). The DT employed an 80%/20% split for training and testing data. Additionally, a radar plot was generated to analyse the importance of each experimental feature. This encompassed factors such as the average diameter of TNTs, cell seeding density on the samples, samples annealed at high temperatures, cell incubation time, and sample sterilisation methods. Li et al. [188] used the SVM algorithm to identify the effects of various parameters on the mechanical behaviours of biodegradable magnesium (Mg) implants, which include metal forming processes and procedural temperature (Fig. 4d).
Applications of data mining/processing in biomaterials. a Unsupervised self-organising map (SOM). Reproduced with permission. Copyright 2019, Elsevier [177]. b K-means clustering method to analyse bioceramic micropores. The yellow dots indicate the centres of each cluster. Reproduced with permission. Copyright 2019, Authors [199]. c The decision tree (DT) model for investigating cell proliferation on titanium dioxide nanotubes. Reproduced with permission. Copyright 2021, Authors [189]. d The flowchart uses the support vector machine (SVM) to estimate the mechanical properties. Reproduced with permission. Copyright 2021, Elsevier [188]. e A fully connected neural network to identify polymer configurations (globule, anti-Mackay, Mackay), including input layer, hidden layer, and output layer composed of neurons (circles). Reproduced with permission. Copyright 2017, American Physical Society [178]
It is worth noting that some ML-based studies on material sciences for engineering applications could also be rather useful [218,219,220,221,222,223,224]. For example, Wei et al. [178] employed an ANN model to classify different states of polymeric configurations, in which the classification can offer a novel and intuitive way to unravel the phase transitions between different polymers. Tripathi et al. [198] used the PCA method to filter noise data for identifying micro-damage in piezoelectric ceramics. Chittam et al. [187] investigated the performances of the logistic regression, SVM, and RF algorithms for data mining/processing in Mg-alloy, which presented a detailed framework on how to use these powerful data science tools in development of biomaterials.
3.3 Digital Twins for Biomaterials
Once pattern/feature recognitions on biomaterial datasets are achieved through some ML procedures, how to relate the identified parameters with material properties becomes a critical issue. In this regard, various ML approaches can be used to establish the relationships between identified parameters and desired biomaterial properties, which typically serve as digital twins [225,226,227,228] of their in vivo and/or in vitro experimental counterparts to predict real-time responses with sufficient accuracy when varying different parameters or patterns. The digital twin constructs a solid bridge between a physical biomaterial and its virtual counterpart, enabling to apply computational modelling techniques to accelerate the design process of new biomaterials [229, 230].
A number of studies have attempted to apply ML-based approaches for establishing digital twins for biomaterials in literature [231,232,233,234]. For example, Epa et al. [180] employed a three-layer NN to model the adhesion of human embryonic stem cell embryoid bodies (hEB) on the various polymeric surfaces. Rostam et al. [182] modelled the immune response of cells to polymer surfaces by using the RF, SVM, and NN models, offering a potential tool for the “immune-instructive” rational design of polymers. Vassey et al. [184] employed the gradient boosting regression method [235] to correlate structure-surface with cell-response, paving a futuristic way to modulate inflammatory responses by rational design of biomaterials as shown in Fig. 5a. The gradient-boosting regression model, trained on a dataset, predicted cell attachment and phenotype based on various surface features. The attachment of macrophages, categorised as high (blue), medium (green), or low (orange), was correlated with the sizes of topographical features in terms of total pattern area (μm2). Robles-Bykbaev et al. [181] investigated the osteocyte growth in scaffolds composed of type I collagen, in which the linear and nonlinear logistic regression models were used to simulate the degradation of collagen together with osteocyte growth and stem cell growth (Fig. 5b). The supervised RF algorithm classified the training samples into four distinct maps. The statistical learning model encompassed both linear/nonlinear regression and Long Short-Term Memory (LSTM) neural networks to predict biological activities. The proposed digital twins exhibited fairly promising results for estimating the biodegradation of collagen scaffolds through image data, which could serve as a potential tool for scaffold design. Burroughs et al. [179] used the RF to model cellular responses to topographically-patterned microscopic polymers, which provides an alternative numerical strategy in design of biomaterials for regenerative medicine (Fig. 5c). In Fig. 5c, the ChemoTopoChip layout features the colours representing various chemistries. The scatter plots illustrate (i) human immortalised mesenchymal stem cell (hiMSC) alkaline phosphatase intensity using a RF model with indicator variables for chemistries and topographical descriptors, and (ii) human macrophage polarisation using a RF model with indicator variables for chemistries and topographical descriptors. The line \(y=x\) represents an ideal fit, and \(R2\) corresponds to the goodness of fit.
Applications of digital twins for biomaterials. a Machine learning predicting structure-surface with cell-response. Reproduced with permission. Copyright 2020, Authors [184]. b ML modelling of cellular growth and degradation of collagen. Reproduced with permission. Copyright 2019, Authors [181]. c Chemical and topographical features enhancing the responses of both cell types using ML. Reproduced with permission. Copyright 2021, Authors [179]. d ML prediction of drug release. Reproduced with permission. Copyright 2022, Royal Society of Chemistry [183]
Concerning biomaterials for drug delivery applications, Santana et al. [183] integrated a perturbation theory with ML approaches for predicting biological responses (e.g., probability of drug deviation from an anticipated dose) to nanoparticles that were designed for drug release. The study compared a number of regression models, including logistic regression, DT, NB, RF, and ANN models. The results, as depicted in Fig. 5d, revealed that the RF and NN models outperformed the others in terms of predictive accuracy. The investigation considered different drug release systems: No. 1, pristine nanoparticles with linked drugs; No. 2, coated nanoparticles with drugs linked to the nanoparticles; and No. 3, coated nanoparticles with drugs linked to coating agents. A combination of the trained perturbation theory and ML models was employed to predict the success of drug release systems using various molecular and coating descriptors.
In the material engineering field, substantial efforts have been made to apply various ML techniques for predicting or modelling mechanical and/or physical properties of materials, which can also be applied to design of biomaterials [236,237,238]. Typically, ML approaches can be used to predict polymeric material properties such as dielectric constant, glass transition temperatures, and bandgap, which are some important clues for biological responses [185]. Several studies have been reported to characterise mechanical and physical properties by ML techniques [190, 191, 193, 194], demonstrating great potential for biomedical applications. For example, Moghadam et al. [192] employed an ANN model to predict the bulk modulus of metal-organic materials, which enabled to establish structural-mechanical stability for 3358 base-materials with starkly different morphologies. Yang et al. [207] used a deep-learning CNN model to construct a digital twin for evaluating the stiffness of composite with different base materials. Various digital twins for ceramics were also widely reported [200,201,202,203], which are expected to be applied for future studies in biomedical engineering. Table 2 summarises more studies applying ML approaches in modelling material properties for the reference of biomaterial applications.
3.4 Data-Driven and Machine Learning-Based Design for Biomaterials
ML approaches in data mining/processing enable to identify key patterns and/or parameters for constructing proper digital twins, which can be used for attaining desired material properties effectively. In literature, numerous studies have adopted different ML techniques for devising novel biomaterials. For example, Damiati et al. [186] explored the optimal design of a biodegradable polymer for drug delivery application. In their study, the drug delivery vehicles were based upon Poly (D, L-lactide-co-glycolide) (PLGA), where a non-steroidal anti-inflammatory drug (NSAID) Indomethacin (IND) was loaded. A multi-layer ANN model was employed to predict PLGA droplet sizes with respect to the input of PLGA concentration (Fig. 6a) and flow rates of both PLGA and aqueous phases, and thus the desired polymer particles can be tuned using the ANN model. Wu et al. [197] investigated the design of a titanium alloy with the desired Young’s modulus close to that of human bone. The study involved several key steps, including property prediction using two ANNs, mass spectrometry (MS) temperature filtering, and plotting combined maps, etc. These two ANN models were established to predict the martensitic transformation temperature (i.e., MS temperature) and the resulting Young’s modulus of the Ti-alloy, as illustrated in Fig. 6b. The constituents of the Ti-alloys, namely Ti, Nb, Zr, Sn, Mo, and Ta, were considered as the inputs. Using the ANN models, six groups of Ti-alloys were obtained, with Young’s modulus ranging from 40 to 65 GPa, validated through the dedicated experimental testing.
Applications of machine learning (ML) in design for biomaterials. a Correlation between the observed and predicted PLGA droplet diameter. Reproduced with permission. Copyright 2021, Authors [186]. b Illustration of the operational process of βLow-assisted alloy design. Reproduced with permission. Copyright 2020, Elsevier [197]. c ML design of high-entropy alloys. Reproduced with permission. Copyright 2019, Elsevier [196]. d Microstructures of synthesised materials designed by ML. Reproduced with permission. Copyright 2020, Authors [204]. e ML design of hydroxyapatite nanopowders as bone fillers. Reproduced with permission. Copyright 2021, Elsevier [262]. f Hierarchical design construction and ML applicability for stronger and tougher microstructural materials. Reproduced with permission. Copyright 2018, Authors [209]
Wen et al. [196] conducted an ML-based design of high entropy alloys with a great hardness, as illustrated in Fig. 6c. The alloys were derived from the Al-Co-Cr-Cu-Fe–Ni HEA system. An experimental dataset containing material compositions and physical properties was utilised to train a SVM model for predicting the hardness. Iteration loop I involved constructing a ML-based surrogate model (\({y}_{i}=f({c}_{i})\)) with a training dataset, which was then applied to a search space to predict the properties and associated uncertainties. A utility function for Design of Experiment (DOE) was employed to select a candidate by balancing the exploitation and exploration. After synthesising and measuring the recommended candidates, the new data were incorporated into the training dataset, facilitating iterative improvement of the surrogate model. Iteration loop II was basically similar to Iteration loop I, with the introduction of a feature pool. In this loop, a ML-based surrogate model was trained from compositions (\({c}_{i}\)) and preselected physical features (\({p}_{i})\), denoted as \({y}_{i}=f({c}_{i},{p}_{i})\). In their study, seventeen new alloys were optimised with higher hardness than the training dataset, showcasing a potential framework for tailoring the mechanical properties of other metallic alloys.
In design of bioceramics, similar strategies have also been taken to explore high-entropy ceramics [261]. For example, Kaufmann et al. [204] employed the RF method to explore the entropy-forming ability of disordered metal carbides. A set of material features reflecting essential chemistry, physics, and thermodynamics of each constituent was taken as inputs (Fig. 6d). In Fig. 6d, the first column presents an electron micrograph for each of the synthesised compositions. Columns 2–6 display the selected Energy Dispersive X-ray Spectroscopy (EDS) chemistry maps for each of the five metal cations present in every system. Column 7 is an electron backscatter diffraction (EBSD) map of the grain structure, revealing the effect on grain size in multi-phase compared to single-phase compositions. Compositions are listed from the largest to the smallest ML predicted entropy-forming ability (scale bar 100 µm). Yu et al. [262] investigated the structural behaviour of nanosized substituted hydroxyapatite (HA) powders using different ML techniques, in which a multi-layer perceptron (MLP) was adopted to model structural characteristics, and a genetic programming (GP) technique was employed to appraise the strength of the predictive model (Fig. 6e). Note that the ANN inputs the chemical compositions and outputs crystallite size (D), micro strain (η), and grain boundary volume fraction (f) of the various substituted HA nanopowders for design.
Design of biocomposites is commonly associated with a large design space, thus becoming a fairly demanding yet an active research field favouring some ML techniques [208, 211, 263]. For example, Gu et al. [209] systematically studied the design of bioinspired hierarchical composites using the ML techniques, as shown in Fig. 6f. The microstructure comprises a detailed assemblage of unit cells, which are then converted into a data matrix of building blocks encoding the individual unit cells. Strength and toughness ratios of designs were computed from the training data and ML output designs. Strain field plots were obtained from digital image correlation (DIC) measurement for ML optimisation. An ANN model was employed to predict the toughness and strength by taking material components in different locations of a unit cell as inputs. The new microstructural patterns obtained from the ANN model have exhibited a higher toughness and higher strength, the design of which was further prototyped by using AM techniques and validated by the experimental tests. Han et al. [210] proposed a ML framework for the design of bioactive glass used for biomedical applications. To precisely predict the dissolution behaviour of the bioglass composites, they compared the performances of several typical ML-based regression models, such as hybridising the RF model with the additive regression (AR-RF), SVM, ANN, linear regression, and Gaussian process regression models, where the AR-RF had proven to be of better performance over the other models. The proposed ML model could be used to design new bioglass composites with a controlled release and is expected to form a useful tool for considering other physical, chemical, biological, and mechanical properties. Table 3 summarises some more recent studies for the data-driven and ML-based designs of materials/biomaterials.
4 Machine Learning in Biomechanics and Mechanobiology
Biomechanics plays a significant role in biomedical engineering, addressing a diverse array of healthcare objectives spanning from the body level to tissue and cellular levels [284, 285]. Mechanobiology signifies an emerging field that also investigates physical forces and mechanical properties of biological systems but focuses more on their spatial-temporal effects on regulating cellular/tissue activities. Recently, ML approaches have demonstrated their efficacy in the realms of biomechanics and mechanobiology, tackling the intricate knowledge required for these interdisciplinary features [36, 95, 286,287,288,289,290]. The following subsections detail the state-of-the-art developments in these fields and explore the computational strategies employed in multiscale modelling. It should be noted that in this review, the relevant studies were identified using such keywords as “machine learning” or “data-driven,” and combined with biomechanics, mechanobiology, and multiscale modelling in Web of Science Core Collection.
4.1 Biomechanics
4.1.1 Body Movements
ML techniques have been increasingly used to study biomechanics in the aspect of body movements, especially combining with signals acquired from various wearable and other devices. For example, several studies have showcased the applications of ML algorithms in analysing knee-specific biomechanics in conjunction with inertia sensors [83, 95, 291,292,293,294,295,296]. These studies focused on recording peak tibial acceleration, which exhibited a certain correlation with vertical ground reaction force (vGRF), knee flexion angle (KFA), knee extension moment (KEM), and sagittal plane knee power absorption (KPA) (Fig. 7a–c). For instance, Fig. 7c illustrates the model consisting of seven rigid segments and 16 Hill-type muscles (blue) with seven virtual inertial sensors (red): namely 1-iliopsoas, 2-glutei, 3-hamstrings, 4-rectus femoris, 5-vasti, 6-gastrocnemius, 7-soleus, and 8-tibialis anterior. In these biomechanical studies, the ML regression models (e.g., ANN, linear and nonlinear regression) could predict either global vGRF or knee-specific measures (e.g., KFA, KEM, KPA) by extracting various features such as shank and foot angle, running speed, ground slope (Fig. 7d). In general, the acceleration signals of a step are transformed into a feature vector representation, and a structured prediction algorithm enables to map the sequence of input vectors to the most likely gait segmentation sequence.
Applications of machine learning (ML) in the measurement of body movements. a Sensors positioned on the lateral aspect of the torso, upper arm, forearm, and hand. Reproduced with permission. Copyright 2020, Authors [83]. b Marker placement for standard biomechanical gait analysis. Reproduced with permission. Copyright 2018, Authors [95]. c Conceptual drawing of a musculoskeletal model. Reproduced with permission. Copyright 2020, Authors [294]. d ML applied to the signals. Reproduced with permission. Copyright 2021, Elsevier [293]. e Alignment of the 3D triad to the images for trunk orientation and matching of the 3D scapula model to the images for scapula orientation. Reproduced with permission. Copyright 2019, Elsevier [297]
A similar strategy has also been reported for scapular kinematics [297], where humeral orientations and acromion process positions obtained from the motion capture data were used to train a multi-layer ANN model for the estimation of scapula orientation (Fig. 7e). Table 4 provides a summary of the studies using ML in biomechanics for body movements.
4.1.2 Hard Tissue
Biomechanics of hard tissues is critically important for unravelling injuries, diseases, trauma, and design for implantable devices [312, 313]. Hard tissues, typically bones, are less prone to damage but often result in serious consequences when injuries occur [314]. While non-invasive imaging technologies such as X-ray and computed tomography (CT) have been widely used for detecting bone fractures, diagnoses relying on the human experience are often labour-intensive [315]. Moreover, micro-fractures are often challenging to be detected properly due to image ambiguity, noise, and other knowledge-dependent limitations [316]. To overcome this issue, ML approaches have been employed to classify bone fractures using image data [317,318,319,320,321,322,323,324,325]. Deep learning-based CNN and ANN models have proven effective in understanding bone microstructures and their fracture mechanics, as illustrated in Fig. 8a–d. For example, Fig. 8a illustrates a flowchart for three classification cases. Following a semi-automated cropping phase, a classic CNN was used as a baseline for classification, characterised by subsequent binary networks. The class activation map was then employed to visualise where the network was focusing. Instead of directly using images, Villamor et al. [326] established patient-specific FE models based on Dual-Energy X-ray absorptiometry; subsequently, the data extracted from finite element (FE) analyses, together with clinical information, were used to train the SVM capable of classifying potential hip fractures (Fig. 8e).
Machine learning (ML) methods for detecting bone fractures. a Flowchart illustrating the three classification cases. Reproduced with permission. Copyright 2020, Elsevier [319]. b Optimised convolutional neural network (CNN) structures for the classification of cortical bone images and trabecular (cancellous) bone images. Reproduced with permission. Copyright 2021, Elsevier [317]. c Gradient-weighted Class Activation Mapping (Grad-CAM) activation heatmaps for the optimised CNN on pristine and failed cortical bone images, with the heatmaps overlaid on the original images. Reproduced with permission. Copyright 2021, Elsevier [317]. d Grad-CAM activation heatmaps for the optimised trabecular bone CNN on pristine and failed trabecular bone images, with the heatmaps overlaid on the original images. Reproduced with permission. Copyright 2021, Elsevier [317]. e Input parameters for predicting fracture risk, including geometry, bone density, boundary conditions, and material properties. Reproduced with permission. Copyright 2020, Elsevier [326]
Apart from bone fracture identification, ML approaches have also been widely employed for bone mechanics studies. Some early works can be traced back to 2004 when Lucchinetti et al. [327] employed an ANN model to inversely identify Young’s modulus and Poisson’s ratio of a small trabecular bone from experimental tests. Recently, Nazemi et al. [328] employed an ANN model to characterise the bone density-modulus relationships by correlating the stiffness of cortical and trabecular bone between the FE-based results and experimental tests. The comparisons of density-modulus relationships between the ANN-derived and literature are illustrated in Fig. 9a. Vukicevic et al. [329] used an evolutionarily assembled ANN model to predict bone fracture resistance curves under different age groups. As illustrated in Fig. 9b, the specimens from cortical bone underwent compact tension tests to prepare the R-curves with specific values of crack size and stresses. Then, evolutionary assembling of ANN was performed to obtain the age-specific R-curves. Rahmanpanah et al. [330] employed two ANN models to predict the load–displacement curves of a long bone, in which the trained ANN models can successfully predict responses of specific bone samples that were not used in the training process (Fig. 9c). In their study, the experimental tests were first conducted to obtain bone length (\(l\)), load exposure (\(t\)), limb side (\(s\)), horse age (\(l\)), and strain components (\({\varepsilon }_{1},{\varepsilon }_{2}, {\varepsilon }_{3}, {\varepsilon }_{4}, {\varepsilon }_{5}, {\varepsilon }_{6}\)). Then, these variables, along with the applied load that was predicted by the first ANN, comprised the input variables of the second ANN. Mouloodi et al. [331] employed the ANN as a regression model to predict bone stiffness under compressive cyclic loading, in which the applied force, exposure time, bone anatomy, and age were used as input variables and exhibited a fairly accurate prediction compared with the experimental results (Fig. 9d). More studies using ML approaches in bone mechanics are outlined in Table 5 for a reference.
Machine learning (ML) in bone mechanics. a Artificial neural network (ANN)-derived density-modulus relationship for proximal tibial subchondral trabecular and cortical bone along with existing density-modulus relationships in the literature. Reproduced with permission. Copyright 2017, Elsevier [328]. b Prediction of bone fracture resistance curves using ML. Reproduced with permission. Copyright 2018, Taylor & Francis [329]. c ML model for predicting long bone load–displacement curves. Reproduced with permission. Copyright 2020, Elsevier [330]. d ML predicting bone stiffness. Reproduced with permission. Copyright 2021, Taylor & Francis [331]
4.1.3 Soft Tissue
Soft tissues play a major role in connecting, supporting, and stabilising hard tissues and organs. Investigating soft tissues poses challenges due to their intricate nonlinear properties arising from heterogeneous and multiphasic microstructures, for which conventional methods may become inefficient and less effective [23, 287, 350, 351]. To tackle these challenges, promising solutions using ML techniques have gained particular interest from research communities recently.
For example, ML approaches have been utilised as surrogate models to predict stress distributions in arterial walls, which is crucial for estimating the rupture risks of atherosclerotic plaques [352, 353]. Specifically, the FE analyses were conducted to derive stress distributions in arterial walls. The resulting FE data served as labelled ground truth data to train the ML models, such as SVM [352] (Fig. 10a) or CNN [353], with the inputs encompassing geometric parameters and arterial pressure. In Fig. 10a, the colour-coded surfaces correspond to the predicted pressure risk ratios. The results underscore the potential of ML methods for real-time diagnosis directly utilising clinical data, thereby reducing or eliminating the need for extensive FE analyses.
Machine learning (ML) in soft tissue mechanics. a Support vector regression using shape feature parameters. Reproduced with permission. Copyright 2017, Springer Nature [352]. b The ML model predicts the zero-pressure shape of the aorta. Reproduced with permission. Copyright 2018, John Wiley and Sons [354]. c The comparison between the SVM model and the ANN model to predict tissue deformation (magnitude errors). Reproduced with permission. Copyright 2017, Elsevier [355]. d The schematic of the ML approach to predicting elastic properties of collagenous tissue through images. Reproduced with permission. Copyright 2017, Elsevier [356]
Liang et al. [354] presented an inverse problem to predict the geometry of a zero-pressure aorta using a multi-layer ANN model (Fig. 10b). The approach parameterised the input shapes as sets of scalar values, named as shape codes. These shape codes were then transformed from input to output shapes through a nonlinear mapping, ultimately decoding into the zero-pressure shape. Similar methodologies have been applied to predict real-time tissue deformation by using the trained ML models such as ANN and SVM [355]. Ground truth data were obtained using the patient-specific FE models subjected to various boundary conditions, altering the magnitude or position of applied external forces (Fig. 10c).
In the realm of soft tissue modelling, ML-based approaches have been instrumental in developing frameworks to predict the elastic properties and nonlinear anisotropic strain–stress curves of collagen tissues [356]. As shown in Fig. 10d, a PCA model was used to parameterise the equal-biaxial stress–strain curves and quantify the overall stiffness. Subsequently, a customised CNN model extracted the structural patterns of collagen tissues and identified overall stiffness for classification purposes. Concurrently, the CNN model was used to perform the regression to predict the PCA parameters. Given the limited experimental data available for soft tissues, an unsupervised ML method combined with supervised learning and data augmentation was proposed to overcome this challenge effectively. Furthermore, Nguyen-Le et al. [357] introduced a novel deep learning framework to predict Pelvis soft tissue deformation. The FE analyses generated a simulation-based database for training and testing. LSTM neural network and deep neural network effectively handled high-frequency oscillation signals, demonstrating better accuracy in predicting soft tissue deformation in real time. Dalton et al. [358] proposed a ML-based framework for modelling soft tissue mechanics, leveraging a Graph Neural Network (GNN) trained in a physics-informed manner to minimise a potential energy function. This framework accommodates unique soft-tissue geometries of individual patients, thus enhancing computational efficiency and accuracy by avoiding low-order approximations.
ML approaches have also been instrumental in estimating constitutive parameters of cardiovascular tissues. For example, Cilla et al. [359] demonstrated the possibility of using ML techniques to fit the constitutive models based on a strain energy function, in which the nonlinear strain–stress curves obtained from the uniaxial experimental tests were used to train an ANN model which could substantially reduce computational costs. Liu et al. [360] successfully used a multi-layer ANN model to estimate constitutive parameters in a strain-invariant-based fibre-reinforced hyperelastic model [361], which was customised for aortic walls. In their study, the training data were generated by the FE simulations with pre-defined nonlinear and anisotropic elastic properties [362, 363]. Then, the systolic and diastolic deformations subject to two starkly different blood pressures were input to the ANN model, which comprised of an unsupervised shape encoding module and a supervised nonlinear mapping module to overcome the issue of data inefficiency, thereby enabling to identify in vivo material parameters in a time-efficient fashion. Gonzales-Saiz et al. [364] developed an ML-based framework to model complex and nonlinear material behaviours of soft tissue. They presented a multiphysics model-driven framework to optimally select the suitable model kinematics, rheological components, and their combination for a given set of experimental curves.
4.2 Mechanobiology
Mechanobiology aims to investigate mechanical effects on tissue or cellular behaviours, which can be traced back to the Wolff’s law for bone remodelling in response to mechanical forces in the 19th Century [365]. Due to recent advances in experimental tools and novel methods, in vivo mechanical forces can be accurately measured [366], providing meaningful ways to gain deep insights into how tissues and cells interact with their physical surroundings, thereby facilitating the development of future treatments from a mechanobiological perspective.
Notably, bony tissue plays a crucial role in safeguarding vital organs such as the brain, heart, and lungs, as well as contributing to load transfer and facilitating proper body movement [367]. The adaptive remodelling and regeneration of bony tissues are paramount for understanding such critical processes as bone healing, distraction [368], and its interaction with prosthetic devices [369]. Consequently, numerous studies have delved into mechanobiological modelling at various levels—organs, tissues, and cells [25, 37, 370]. In tissue scaffold engineering, for instance, dynamic bone growth outcomes have been demonstrated to be significantly important for bone scaffold design [34, 38, 371,372,373,374,375]. Lured by this promise, a number of significant studies have investigated tissue ingrowth in scaffolds based on different mechanobiological models [33, 34, 233, 371, 376,377,378,379,380,381,382,383,384,385,386,387,388], as summarised in Table 6.
The critical importance of bone osseointegration and remodelling around implants lies in their potential performance for either successful implantation or failure/loosening of positioned devices. In this regard, Ghosh et al. [389] carried out a design optimisation of implant surfaces to enhance osseointegration and bone growth, in which three different parameterised surface models were employed as design candidates. To reduce computational cost for evaluating bone growth during the optimisation process, a NN model was employed as a surrogate. As bone growth results could be evaluated in a real-time fashion, design optimisation can be performed using GA and the NN model without the need for time-consuming time-dependent FE analyses.
In effect, mechanobiology also plays a key role in depicting the behaviour of soft tissues and their associated diseases. Numerous studies are currently underway to explore biomechanical forces on vessels in vivo or in silico [390,391,392]. At a cellular level, mechanosensation and mechanotransduction can help understand cell proliferation and differentiation in response to mechanics [393,394,395,396,397,398,399].
Despite the crucial roles of mechanobiology in cells and tissues, there have been relatively fewer studies utilising ML techniques in this area compared to generic biomechanics. In this regard, Dattatrey et al. [400] employed an ANN model to predict bone remodelling parameters in response to patient-specific loads, offering a potential for regulating the neobone responses. Tiwari et al. [401] employed an ANN model to establish the relationship between the loading parameters (strain magnitude, frequency, and cycle) and bone remodelling parameters, intending to create a comprehensive in silico model for predicting bone remodelling under varying load conditions.
For regulating cellular behaviour, Bonnevie et al. [402] applied the ML approaches to investigate the mechanosensation of cells in heterogeneous microenvironments. In their study, a SOM model was employed to reduce dimensionality of cellular shapes, and a multi-layer ANN model to predict the mechanobiological state of Yes-associated protein and transcriptional coactivator with PDZ-binding motif with different cell morphology, which was further used to identify contractile cell populations distinctly.
4.3 Multiscale Modelling
Biological tissues exhibit intricate and often highly nonlinear and anisotropic material behaviours. To develop high-fidelity computational approaches, various typical constitutive models for tissue mechanics, such as hyperelastic [403], viscoelastic [404], and poroelastic [405] models, have been explored. The accuracy of these different constitutive models normally relies on correlating their model parameters with experimental data. Over the past decades, both hard and soft tissues have been identified to possess significant heterogeneity at the microscopic level [406, 407]. This recognition addresses the challenge of why a single constitutive model often falls short in adequately matching experimental results without sacrificing generality.
To overcome this difficulty, various multiscale finite element (FE2) techniques have been developed for modelling heterogeneous tissues, which are summarised in Table 7. Conventional multiscale finite element (FE2) methods commonly employ a homogenisation approach [408] to generate effective constitutive models for macro-FE analyses. However, the complex microstructures of biological tissues are randomly distributed, making the homogenisation process computationally expensive [348]. Extracting a single representative volume element (RVE) that characterises all heterogeneous microstructures and material behaviours can be demanding, if not impossible, necessitating further studies [409].
For example, structures of tissue scaffolds play a crucial role in new bone regeneration, influencing the mechanical stimulation that regulates cell proliferation and differentiation [410]. Therefore, computational methods (in silico) have been introduced as an effective alternative to time-consuming and expensive in vitro and in vivo experiments [411,412,413,414,415]. Nevertheless, these scenarios necessitate multi-FE analyses at microscale. Particularly for design optimisation and inverse determination of scaffolding parameters, the computational costs of conventional FE2 methods can become prohibitive, severely limiting their practical applications [409].
To overcome such computational hurdle, several promising ML studies have taken a significant step toward addressing the challenges imposed by conventional FE2 methods. In bone remodelling studies, an ANN model was applied at the microscale level. Here, the nominal stress derived from macro-FE analyses and cyclic loading frequency served as inputs to predict the average bone density, effective damage, and bulk Young’s modulus of each representative volume element for macro-FE bone remodelling analyses (Fig. 11a) [441, 442]. In more detail, a whole bone was analysed using a macro-FE model. At the microscopic level, a micro-FE model was developed using a trained NN incorporated into the FE code Abaqus via a user subroutine UMAT. During the FE calculation at the macro level, the NN was invoked at every integration point to compute the averaged outputs representing the RVE behaviour at the mesoscale.
Machine learning (ML) in multiscale mechanobiological modelling. a Multiscale approach for bone adaptation simulation using combined finite element (FE) and artificial neural network (ANN) computation. Reproduced with permission. Copyright 2011, Elsevier [441]. b ML in predicting multiscale bone regeneration outcome in bone scaffolds. Reproduced with permission. Copyright 2021, Springer Nature [409]
Regarding mechanobiology-driven bone ingrowth in tissue scaffolds, Wu et al. [33] developed an ML-based multiscale computational framework to predict bone ingrowth in bulk scaffolds, as shown in Fig. 11b. In stage I, the in vivo image data were correlated with the ML-based in silico results to inversely determine the subject-specific parameters (R1, R2, R3, K, s) associated with a mechanobiological remodelling algorithm. NN-3 outputs the Pearson correlation coefficients (P1, P2) between the in vivo and in silico data. In stage II, microscopic bone densities within the scaffold’s representative volume element (RVE) were input to NN-2 to evaluate the strain energy density (SED). This SED served as a mechanical stimulus to update bone densities using the Wolff’s law model established with a set of inversely identified algorithmic parameters for properly predicting bone formation. In stage III, homogenisation of the RVE was performed using NN-1 to predict effective mechanical properties. Given the heterogeneous nature of tissue ingrowth within a scaffold, characterised by spatial–temporal variations in mechanical stimuli, the ML techniques were employed at the microscopic level. Here, homogenisation and strain energy density of RVEs were directly derived using the ANN models instead of time-consuming micro-FE analyses (Fig. 11b-ii and b-iii). It is noteworthy that the developed ML-based multiscale framework significantly reduces computational costs and provides new opportunities for both inverse determination of different bone ingrowth parameters and design optimisation studies that might be prohibitive when using the conventional FE2 approaches.
Regarding the multiscale modelling of tissue mechanics, Pled et al. [443] proposed a ML-based approach for an inverse problem, in which an ANN model was employed to output the identified effective elastic properties of heterogeneous microstructures, as shown in Fig. 12a-i. In their study, the heterogeneous microstructure with random compliance field \({\varvec{S}}\) was parameterised by four hyperparameters \({H}_{1}, {H}_{2}, {H}_{3}, {H}_{4}\) that represent the dispersion parameter, spatial correlation length, mean bulk modulus and mean shear modulus, respectively (Fig. 12a-ii). These four parameters can recover any random compliance field \({\varvec{S}}\) by a nonlinear mapping (Fig. 12a-iii). The region of interest (ROI) for a given domain was analysed by both the High Fidelity Computational Mechanical Model and Computational Homogenisation Model with nine statistic-dependent parameters (\({Q}_{1}\) to \({Q}_{9}\)) that were taken as the inputs for the ANN model to output four apparent elastic properties of a given compliance field \({\varvec{S}}\) (Fig. 12a-iv). The developed approach was applied in a numerical example based on the experimental data from real heterogeneous bovine cortical bone, which exhibits promising results in terms of both modelling accuracy and computational efficiency.
Machine learning (ML) in multiscale modelling of tissue/organ biomechanics. a A statistical inverse problem in multiscale biomechanics. Reproduced with permission. Copyright 2020, Elsevier [443]. b The multiscale modelling of liver using a ML algorithm. Reproduced with permission. Copyright 2019, Springer Nature [444]. c A multiscale data-driven approach for bone tissue biomechanics. Multiscale data-driven results. Top: Macroscopic strain field. Bottom: Microscopic strain field in the RVE for the highlighted point of the macroscale. Left: longitudinal strain component. Right: transversal strain. Reproduced with permission. Copyright 2020, Elsevier [445]
Hashemi et al. [444] proposed a ML-based multiscale computational framework for modelling heterogeneous liver tissue composed of versatile shapes of vessels and surrounding soft tissues with anisotropic properties, as illustrated in Fig. 12b. The geometric size of the liver is usually substantially larger than that of its vascularisation, which hinders proper use of a mono-scale FE model that requires extremely fine mesh in some regions with vasculature. Homogenised models with relatively coarse mesh subject to patient-specific geometry then serve as practical solutions, where the homogenised hyperelastic properties of arbitrary heterogeneous microstructures could be determined by a FE framework [444]. Nevertheless, the tedious conventional FE framework may need to be established for every patient-specific model at each ROI, which severely limits its use. To reduce the computational cost and improve the generality, an ANN model trained by the observed data was employed to directly determine the parameters in the Holzapfel-Gasser-Ogden model for anisotropic hyperelastic material, where the inputs were the spatial orientation and diameters of vessels in ROIs. The vascularised liver tissue was discretised into segments, and the geometric parameters of these segments were input to an ANN model to output the homogenised material properties for macro-FE modelling. Compared to the conventional FE2 framework, the error of ML-based results is at an acceptable level but with a substantially lower computational cost.
Homogenisation of microstructures normally relies on the assumption of certain material constitutive models (e.g., linear elastic, hyperelastic, viscoelastic). Nevertheless, even a highly sophisticated constitutive model may fail or not be accurate enough to describe the mechanical properties of a heterogenous segment at the microscopic level. To deal with this issue, Mora-Macias et al. [445] developed a data-driven multiscale approach for the mechanical modelling of heterogenous bone, as shown in Fig. 12c. In the proposed algorithm, a biaxial compressive test was carried out on a cortical bone sample. A digital image correlation (DIC) system was used to record the strain field in the course of loading. For a given microscopic ROI as a RVE, the homogenised macro strain \({{\varvec{\upvarepsilon}}}^{M}\) can be obtained using the micro strain \({{\varvec{\upvarepsilon}}}^{m}\) measuring by the DIC. As a result, the experimentally observed dataset contains a pair of macro strain and marco stress (\({{\varvec{\upvarepsilon}}}^{M}({{\varvec{\upvarepsilon}}}^{m}), {{\varvec{\upsigma}}}^{M}\)), in which the macro strain \({{\varvec{\upvarepsilon}}}^{M}({{\varvec{\upvarepsilon}}}^{m})\) is composed of the micro strain field information \({{\varvec{\upvarepsilon}}}^{m}\). As only a limited number of data could be obtained through the experimental tests, a data-completion technique was proposed to enrich the dataset (\({{\varvec{\upvarepsilon}}}^{M}({{\varvec{\upvarepsilon}}}^{m}), {{\varvec{\upsigma}}}^{M}\)) at both the macroscopic and microscopic levels. A data-driven modelling approach as an extension of the algorithm presented by Kirchdoerfer and Ortiz [446, 447] was then performed by searching for the closest stress–strain pair (\({{\varvec{\upvarepsilon}}}^{M*}, {{\varvec{\upsigma}}}^{M*})\) in the enriched dataset (\({{\varvec{\upvarepsilon}}}^{M}({{\varvec{\upvarepsilon}}}^{m}), {{\varvec{\upsigma}}}^{M}\)). In the proposed data-driven approach, no constitutive model at the microscopic level was pre-assumed. The relationship between the micro strain and the macro strain was directly obtained from an experimental dataset, thereby avoiding the time-consuming homogenisation analyses and thus improving the accuracy of multiscale results at the same time. In contrast to the conventional linear orthotropic material behaviour of cortical bone tissue, they identified the non-smooth patterns of macro stress and macro strain using the proposed data-driven algorithm, demonstrating a potential improvement of a pre-assumed, well-defined model by a data-driven approach.
5 Machine Learning in Biofabrication
Additive manufacturing (AM) technologies pave a relatively new and solid way for novel biofabrication. The advent of ML techniques has generated a substantial impact on biomedical engineering without the exception in biofabrication due to their distinguished benefits and exceptional flexibility for patient-specific strategies [448,449,450,451,452]. This section aims to comprehensively review the ML techniques involved in biofabrication, focusing on the four crucial aspects: structural design, metamaterial development, optimisation of processing parameters, and in situ monitoring of manufacturing. As general AM techniques have been well developed, readers who are interested in their other generic applications than the biomedical fields can refer to some more comprehensive review articles [53, 453,454,455,456]. The relevant studies discussed in this section were identified using keywords such as “machine learning” or “data-driven,” combined with structural optimisation, topology optimisation, metamaterial design, processing parameters, and in situ monitoring of AM in the Web of Science Core Collection by 2023.
5.1 Structural Design
Additive manufacturing offers a new technological paradigm and exhibits remarkable advantages over conventional fabrication processes for producing geometrically sophisticated products with desired structural characteristics and functions. This capability bridges the gap between novel design and effective realisation [457, 458]. Structural design signifies a crucial step in the workflow of biofabrication, often involving iterative trial-and-error processes and various experimental tests that may not be available with ease [459]. As a result, the so-called rational design has drawn particular interest from the research community and industry sector [460, 461], in which one needs to know how the structural change can affect desired functionalities so as to reasonably tune structural design with an acceptable cost. Nevertheless, the largely improved design freedom offered by AM may increase the cost of conventional rational designs. To this end, ML techniques have been introduced to shed new light on the conventional design workflow [462,463,464].
5.1.1 Parametric Optimisation
One of the most commonly used rational design methods is parametric optimisation [465, 466]. In contrast to trial-and-error approaches, desired structural performances are treated as objectives and/or constraints with respect to design variables that parameterise the structural configuration, materials and dimensions in an explicit form. Taking design of orthopaedic devices as an example, different types of cellular structures have been comprehensively investigated, such as strut-based lattices [467,468,469], triply periodic minimal surface (TPMS)-based lattices [470], bio-inspired lattices [471,472,473,474], auxetic structures [475], honeycomb structures [476], and origami-inspired structures [477] (as shown in Fig. 13a). After generation of these parameterised cellular structures, computational modelling techniques can be involved to calculate the responses of objectives and constraints, where the ML techniques, as reviewed in Sect. 4, have substantial potential to play active roles, e.g., prediction of bone remodelling [33], interactions with the host environment [389, 402], etc.
Machine learning (ML) in parametric optimisation. a Different types of cellular structures, including strut-based lattices (Reproduced with permission. Copyright 2019, Elsevier [468]), TPMS-based lattices (Reproduced with permission. Copyright 2018, American Chemical Society [470]), bio-inspired structures (Reproduced with permission. Copyright 2020, Elsevier [472]), auxetic structures (Reproduced with permission. Copyright 2019, Authors [475]), Honeycomb-like structures (Reproduced with permission. Copyright 2014, John Wiley and Sons [476]), and Origami-inspired structures (Reproduced with permission. Copyright 2-16, Elsevier [477]). b ML design of implants targeting desired strain pattern. Reproduced with permission. Copyright 2019, John Wiley and Sons [480]. c ML-based design of patient-specific dental implants. Reproduced with permission. Copyright 2018, Elsevier [481]
In parametric optimisation, surrogate models [478] are often employed to approximate the responses of objectives and constraints with respect to design parameters. In this regard, supervised ML techniques show great benefits in parametric optimisation, which vary from the conventional surrogate techniques, such as the response surface model, Kriging model, and radial basis function model [478], to more prevailing ML regression approaches. In literature, there have been several studies [479,480,481] employed NNs as surrogate models to predict the micro strain on bone-implant interfaces, in which the GA was used to optimise the input variables for tweaking the shape, size, and porosity of implants, thereby achieving the desired micro strain to avoid loosening of implants as illustrated in Fig. 13b and c.
Figure 13b-i depicts the internal hollowness of the femoral stems [480]. The ANN model outputs the strain difference, and the GA tunes the input parameters to achieve desired performances and outcomes, including maximum composite desirability (Fig. 13b-iii) and optimum average roof thickness values under different bone conditions (Fig. 13b-iv). In Fig. 13c, the FE analyses were performed to obtain bone-implant interface micro strain and implant stress at different diameters, lengths, and porosities of the porous dental implant for various bone conditions, serving as independent inputs to two ANN models [482]. The desirability function was used as an index to obtain desired micro strain, and the GA optimised these parameters subject to an implant stress constraint.
5.1.2 Topology Optimisation
For rational design of biomedical devices, another widely employed approach is topology optimisation [34, 38, 483]. This technique allows materials to be freely distributed in design domains, facilitating the creation of novel and sophisticated configurations. Topology optimisation enables the exploration of material potential for desired performances by significantly altering structures that notably differ from their initial designs. While intricate geometries generated by topology optimisation were once a bottleneck for fabrication, recent advances in AM technologies have relieved this obstacle, thus largely promoting topological design in real-life applications [484].
In the field of biomedical engineering, topology optimisation has been extensively applied to the design of various implantable devices, including tissue scaffolds [34, 378, 379, 485,486,487,488,489], dental implants [483, 490,491,492,493,494], arterial stents [495,496,497,498], bone fixation plates [38, 499, 500], artificial hips [501, 502], and spinal cages [503, 504]. Conventional topology optimisation methods typically require analytical/numerical sensitivity of objectives and constraints with respect to the design variables (e.g., density-based methods [505,506,507,508,509,510,511] or shape functions (level-set method [512, 513], moving morphable component (MMC) method [514]) to drive topological changes. However, the iterative optimisation process involving repeated FE and sensitivity analyses may not be ideal for achieving real-time responses. To this end, recent studies have coupled ML techniques with conventional topology optimisation techniques, where conventional topological configuration results were used to train ML models for obtaining real-time responses of topology optimisation, as summarised in Table 8.
For example, Yu et al. [515] utilised a CNN-based encoder-decoder network and a conditional generative adversarial network (cGAN) to achieve real-time topology optimisation without any iteration. In this approach, the CNN encoder-decoder predicts some optimal structures subject to different load conditions at low resolution, and then the cGAN takes low-resolution results as input to predict their high-resolution counterparts, as shown in Fig. 14a. Nie et al. [516] employed a generative adversarial network (GAN) to output optimal structures in a real-time fashion, using initial boundary conditions, load magnitude, and desired volume fraction as input images to the GAN model.
Machine learning (ML) in topology optimisation. a Real-time topology optimisation using a convolutional neural network (CNN) and a conditional generative adversarial network (cGAN). Reproduced with permission. Copyright 2018, Springer Nature [515]. b Topology optimisation using a neural network (NN) model. Reproduced with permission. Copyright 2020, Springer Nature [517]. c Multiscale topology optimisation using ANN as a surrogate. Reproduced with permission. Copyright 2019, Elsevier [518]
In addition to achieving real-time topology optimisation, Chandrasekhar et al. [517] used a NN model to parameterise design variables. In this case, a density function in terms of a conventional solid isotropic material with penalisation (SIMP) interpolation model is directly parameterised by the weights and bias associated with the NN model. Thus, the conventional optimisation procedure was modified to train the NN model in terms of the loss function from FE analyses. Once the NN training is finished, the optimisation problem can be solved straightforwardly, as illustrated in Fig. 14b.
Another significant application of ML techniques in topology optimisation is for design of multiscale structures. Many studies have been devoted to various schemes, as outlined in Table 8. Notably, White et al. [518] employed an ANN model as a surrogate to predict the elastic response of microscale unit cells. At a microscale level, the analytical sensitivity of elastic response with respect to the design parameters controlling micro-unit cells was used to train the ANN model, thus enabling the topology optimisation at a macro-scale to use the ANN model for both the elastic responses and sensitivity information, thereby driving the topology optimisation of every single unit cells (Fig. 14c). Note that multiscale topology optimisation can be extensively useful for metamaterial designs that have broad applications in implantable devices.
5.1.3 Manufacturing Constraints
Although AM techniques have demonstrated substantial benefits for fabricating intricate products, the integration of new manufacturing constraints poses significant challenges for real-life structural designs. Notably, the necessity of support structures for large overhang designs in AM [569] and the requirement to avoid self-enclosed cavities in powder-based AM techniques for the removal of residual powder after fabrication [570] are some critical considerations. In the context of metal printing, the impact of thermal residual stress becomes pivotal, as it can lead to crack-induced failure in the fabricated parts [569]. Consequently, the emerging field of design for additive manufacturing, which systematically accounts for these manufacturing constraints, has garnered considerable attention in research communities [571, 572]. Table 9 provides a summary of some noteworthy studies in topology optimisation design for AM, taking into consideration various manufacturing constraints.
To address manufacturing constraints in ML-based topology optimisation, Tan et al. [587] proposed a framework utilising deep learning for design of microstructures. In their approach, a deep learning based Generative Adversarial Network (GAN) model is trained to directly learn geometry constraints from a database. The architecture of the combined model for inverse design is depicted in Fig. 15, where the Generator of Deep Convolutional GAN and CNN are represented by a simple neural network structure for clarity. Figure 15 also shows a flowchart of the inverse design process using the combined model, represented as a simplified version in the grey-coloured box for clarity. In conventional topology optimisation, geometry-related manufacturing constraints such as overhang/self-enclosed holes are typically described by certain numerical models, which may be difficult to use for other geometries generally. In this aspect, their study shed new light on coupling arbitrary geometric constraints by directly training the ML models to learn the key features in an unsupervised fashion, which can be extended to other geometry-related manufacturing constraints with ease.
Machine learning (ML) in topology optimisation coupling with manufacturing constraints. Reproduced with permission. Copyright 2019, Springer Nature [587]
Recently, Weiss et al. [588] proposed a data-driven topology optimisation approach for considering the AM constraints, in which a single fixed minimum feature size and a maximum overhang angle predicted by a data-driven constraint function were integrated with the MMC-based topology optimisation framework. The procedural steps include: (i) setting up the problem with design domain, load, boundary conditions, and an building direction of AM as the inputs, (ii) obtaining a SIMP solution with fixed minimum feature size, (iii) converting the density field into a set of connected morphable components, (iv) adjusting the component endpoints and thicknesses using the MMC algorithm to optimise the objective, (v) assessing the minimum manufacturable thickness for each component using the constraint function, (vi) smoothly reducing density from fully dense to void, projecting each component to the new density field, and (vii) performing FE analysis and sensitivity calculations, with the optimiser updating the decision variables.
5.1.4 Manufacturing Uncertainty
Apart from these manufacturing constraints, AM-induced material and geometric uncertainties are also some crucial parameters to be considered. Material uncertainty may impact the performance of as-built parts, while geometric uncertainty may drastically deviate the geometries of as-built parts from their as-designed counterparts, resulting in considerable variation in functional performance, as reported for development of porous structures [569]. Geometric uncertainty in AM stems from various factors, including: (1) thermal residual stress [589], (2) material shrinkage [590, 591], (3) distortion [592, 593], and (4) manufacturing imperfection [594]. In literature, significant efforts have been dedicated to addressing such critical issues by either minimising geometric uncertainty or simulating the induced sources so as to take into account them in design stage, thereby minimising their impact on their final performance, or ensuring reliability of the built structural systems [580, 595,596,597,598,599,600]. Table 10 summarises some typical studies considering AM-induced uncertainties in topology optimisation.
Nevertheless, the application of ML techniques is still evolving to address these challenges [601,602,603,604]. For example, Ferreira et al. [605] proposed a compensation scheme to reduce deviations between as-built and as-designed parts using ML techniques, in which an automated NN-based modelling method enabled to accurately predict the deviations during AM processes. Huang et al. [606] introduced a Shape Deviation Generator (SDG) as an engineering-informed ML framework that learned from the AM data to control the shape deviations in AM processes.
5.2 Metamaterial Design
In vivo tissue exhibits inhomogeneous structures with notably intricate features ranging from macro to nanoscale. Implantable devices, such as orthopaedic implants, are expected to share similar structural characteristics and material properties with the surrounding host tissues. For instance, an appropriate effective stiffness is crucial for resisting external loads while avoiding severe stress shielding; and an adequate effective permeability ensures cell transformation and nutrition/metabolism transportation. Given these complex and often competing design requirements, metamaterials fabricated using AM techniques are particularly favoured in the design of implantable biomedical devices [621,622,623,624].
Recent studies have introduced innovative approaches to topology optimisation for single-unit cell or periodic structures using data-driven and ML techniques. These approaches offer novel computational tools for designing porous implants with functionally graded microstructures [518, 532, 625,626,627,628,629,630,631]. For example, Wang et al. [626] proposed a multi-response latent-variable Gaussian process (MP-LVGP) for creating microstructural libraries of metamaterials with different classes and sizes. The MP-LVGP model was integrated with multiscale topology optimisation for structures composed of multiple classes of microstructures, which are generally demanding for conventional topology optimisation approaches.
In literature, ML for metamaterial design, combined with AM, has been employed to mimic soft tissue. Chen et al. [632], for example, conducted ML-based design of printable polymers to replicate the mechanical responses of soft tissue under various stress–strain curves. In their study, the functional principal component decomposition approach, coupled with Gaussian process modelling, served as a surrogate model to predict the mechanical responses (stress–strain curves) of simulated metamaterials efficiently. This approach enabled the parametric optimisation for controlling metamaterial structures towards some exceptional material properties. Table 11 summarises the studies using data-driven and ML approaches for metamaterial design, and the structures/morphologies of metamaterials in each study are illustrated in Fig. 16 for clarity and better insights.
Data-driven and machine learning based studies on metamaterial design associated with the index for different structures in Table 11. Reproduced with permission. Copyright 2021, Authors [633]. Reproduced with permission. Copyright 2021, Springer Nature [634]. Reproduced with permission. Copyright 2020, John Wiley and Sons [635]. Reproduced with permission. Copyright 2020, Springer Nature [627]. Reproduced with permission. Copyright 2020, Elsevier [628]. Reproduced with permission. Copyright 2020, Springer Nature [536]. Reproduced with permission. Copyright 2019, John Wiley and Sons [637]. Reproduced with permission. Copyright 2018, American Chemical Society [638]. Reproduced with permission. Copyright 2021, Authors [639]. Reproduced with permission. Copyright 2020, Authors [630]. Reproduced with permission. Copyright 2018, Elsevier [632]. Reproduced with permission. Copyright 2022, Elsevier [640]. Reproduced with permission. Copyright 2020, Authors [642]. Reproduced with permission. Copyright 2019, Authors [643]. Reproduced with permission. Copyright 2023, Authors [645]. Reproduced with permission. Copyright 2023, Elsevier [647]
5.3 Optimisation of Processing Parameters
The processing parameters in AM can significantly impact the properties of as-built parts, particularly when new materials or new AM techniques are employed [453]. While trial-and-error experiments are a common approach to address this issue, they can be inefficient and costly. For this reason, researchers have turned to physics-based modelling to uncover the intricate relationship between processing parameters and the outcome of as-built parts [649,650,651]. Nevertheless, establishing suitable physics-based models for the complex nonlinear relationships involved can be rather demanding as per conventional modelling techniques [454].
To tackle these challenges, ML techniques have been introduced to discern and optimise the relationship between processing parameters and the performance of as-built parts based on specific requirements. The applications of ML techniques in optimising process parameters generally fall into two categories. Firstly, ML techniques developed for data clustering can be utilised to identify qualitative relationships between the processing parameters and as-built outcome/performance, subsequently determining the most crucial parameters for optimisation [651,652,653]. The dataset for this approach can be sourced from existing physics-based models or experimental tests. Secondly, given the intrinsic nonlinear nature of these relationships, regression ML models exhibit significant potential for explicitly quantifying them without relying on physics-based models, enabling the optimisation of process parameters for desired performances. This flexibility allows coverage of different AM techniques, each with its unique set of process parameters [654, 655]. Table 12 provides a summary of typical studies utilising ML approaches to establish the relationships between various processing parameters and critical as-built structural characteristics and properties with different AM techniques.
In the realm of 3D bioprinting, which involves the use of bioink containing live cells for tissue engineering, the significance of processing parameters becomes even more pronounced, affecting both printing accuracy [660] and cell viability [658]. The demand for effective solutions to enhance the stability and precision of cell injection, while simultaneously reducing cell damage, is imperative to the outcome. Take 3D extrusion-based bioprinting (EBB) as an example; here, cell viability is notably influenced by shear stress generated within printing nozzles. This shear stress is, in turn, dependent on such factors as pressure drop and the shape of the nozzles [656]. To tackle this challenge, various ML regression models have been harnessed to predict cell viability, providing valuable guidance for selection of optimal processing parameters [659, 663], as illustrated in Fig. 17a. For inkjet-based bioprinting, a multiobjective optimisation framework has been introduced to address the complexities associated with this technique. In this framework, ANNs play a pivotal role in modelling satellite formation concerning different droplet diameters and speeds [656, 700], as shown in Fig. 17b.
Machine learning (ML) in optimisation of biofabrication processing parameters. a Key components within the Stereolithography-based bioprinting system as well as the 2D patterns of a four-branch vascular structure under printing. Reproduced with permission. Copyright 2020, Springer Nature [659]. b ML-based optimisation of processing parameters in ink-jet-based bioprinting. Four factors (applied voltage, bioink viscosity, surface tension, and nozzle diameter) determine the droplet size and state. Reproduced with permission. Copyright 2019, Authors [700]
5.4 In Situ Monitoring
In situ monitoring of the AM process involves the analysis of various data from multiple tools, such as cameras, X-rays, images, and sensors, to detect and eliminate potential manufacturing defects or undesired properties (e.g., deformation, missing or excess materials, micro-cracks) [701, 702]. In situ monitoring for AM process holds significant promise for automatically analysing real-time signals without laborious human intervention, which enables to provide instantaneous feedback to the AM process, thereby forming a closed-loop control [703].
Data in the AM process is recorded either in a time-series fashion or stored as images [704, 705]. ML techniques for processing time-series signals or computer vision have exhibited considerable potential and superior performances in identifying abnormal changes across various types of information. In this context, many researchers have developed computer vision systems with cameras to acquire image data during the AM process. Trained Convolutional Neural Networks (CNNs) are then utilised to extract features from observed images, such as melt pool, plume, spatter, and droplet patterns [706,707,708,709], as shown in Fig. 18. For example, Fig. 18b outlines the steps to produce a time-integrated image. Each time series of radiographs is segmented and labelled with time, then flattened into a time-integrated image. The extracted features are classified by the CNN models trained with labelled data in different classifications for in situ monitoring of the AM process.
In situ monitor of the additive manufacturing process using machine learning techniques. a In situ monitoring using high-speed and high-energy X-ray imaging techniques in the laser powder bed fusion process. Reproduced with permission Copyright 2018, Elsevier [706]. b In situ X-ray imaging detection of defects. Reproduced with permission. Copyright 2018, Authors [707]. c In situ droplet monitor for liquid metal jet printing. Reproduced with permission. Copyright 2018, Elsevier [708]. d In situ monitor of melt pool in powder-based fusion processes using support vector machine and convolutional neural network models for processing images. Reproduced with permission. Copyright 2018, Elsevier [709]
To form the closed-loop control, ML models can establish relationships between extracted features and real-time printing behaviour (e.g., temperature, speed, voltage), as reviewed in Sect. 5.3. The differences between real-time and desired behaviours can be evaluated, and proper control strategies can automatically adjust processing parameters, forming positive feedback on real-time printing outcomes. Given these promising results, more advanced deep learning frameworks are anticipated to enable in situ monitoring of specific issues occurring in the biofabrication process, thereby enhancing bioprinting to be more robust, reliable, and accurate. Table 13 summarises some further studies on ML techniques for the in situ monitoring of the AM processes, which include their inspection objectives, ML algorithms, AM techniques, and data collection approaches.
6 Applications
6.1 Bone Tissue Scaffolds
Bone loss resulting from trauma, tumours, infections, or injuries poses a significant healthcare challenge, necessitating either temporary or permanent replacement with a suitable functional alternative. Traditional treatment approaches, such as allograft and autograft transplantations, as well as Masquelet procedures, exhibit notable drawbacks, including infection risks, disease transmission, donor site morbidity, and restricted tissue supply [33]. To overcome these limitations, there has been growing interest in synthetic bone scaffolds as a potential solution for addressing bone loss in both research communities and clinical settings (Fig. 19a). These implantable scaffolds are specifically designed to replace bony tissues in vivo, requiring not only sufficient mechanical support but also biologically/chemically inert material properties and the ability to facilitate the transport of nutrients and metabolism crucial for tissue regeneration [34].
Applications of biomedical implants. a Bone scaffold. Reproduced with permission. Copyright 2019, John Wiley and Sons [784]. b Hip implants. Reproduced with permission. Copyright 2017, Authors [752]. c Dental implants. Reproduced with permission. Copyright 2021, Authors [763]. d Coronary stents. Reproduced with permission. Copyright 2016, Elsevier [41]
Table 14 provides a summary of some typical ML-based applications in bone tissue engineering. For example, Entekhabi et al. [743] developed a ML model to predict the degradation rate of a biodegradable scaffold. In their study, the experimental tests were conducted on different samples with varying material constituents. The experimental data were then utilised to train an ANN model for predicting the degradation rate with a fairly low mean squared error (2.68%).
To achieve optimal bone formation in tissue scaffolds, structural scrutiny aligned with subject-specific defect sites is imperative. Consequently, there is a growing demand to efficiently predict bone formation within tissue scaffolds through use of computational modelling techniques, mitigating the need for costly trial-and-error in vivo experimental tests. In this regard, Barrera et al. [744] employed the ML techniques for design of scaffolds, in which a CNN model was employed to directly predict the mechanical properties of innovative scaffolds without experimental tests or numerical models, thereby enabling rapid optimisation of scaffolds for desired biomechanical properties. Moreover, ML techniques have also been applied to guide the bioprinting of bone scaffolds [662, 745, 746].
6.2 Orthopaedic Implants
Hip fractures persist as a significant health concern, particularly in the context of an increasingly ageing population. Consequently, hip replacements utilising artificial implants have become a golden standard for revision surgeries worldwide (Fig. 19b). This widespread adoption underscores the urgency of addressing the challenges associated with such procedures. To this end, several ML-based applications in the realm of hip implants are delineated in Table 14. The introduction of hip implants presents a transformative mechanical environment to be experienced by the host bony tissue. This alteration raises the spectre of possible stress shielding issues, prompting the necessity for patient-specific design of hip implants. Addressing this concern, Cilla et al. [752] proposed a ML-based framework for optimising hip implants. Their approach involved the development of a parametric FE model, quantifying stress shielding effects by assessing the disparity in maximum principal strain between implanted and intact bone models. Subsequently, an artificial neural network (ANN) model, trained from the FE dataset, provided quantified stress shielding values for various parametric models. Such information was then integrated with the GA to determine the optimal parameters conducive to reduction of stress shielding.
Beyond stress shielding, the longevity of hip implants is intricately linked to polyethene wear, a pivotal factor in determining their overall performance. Addressing this concern, Borjali et al. [753] adeptly predicted the polyethene wear rate through the ML models applied to pin-on-disc (PoD) wear experiments. Their innovative approach paved a new avenue for investigating this critical issue by observing diverse operating parameters, potentially mitigating complications such as osteolysis, implant loosening, and mechanical instability.
In clinical applications, ML techniques have found notable utility in the identification of hip implants from plain radiography images owing to their robust performance in imaging analysis [754,755,756,757,758]. This highlights the versatility of ML methodologies in enhancing clinical diagnostics and underscores their potential in augmenting traditional imaging analysis techniques.
6.3 Dental Implants
The prevalence of tooth loss, a consequential issue in oral health, is exacerbated by the ageing population. Apart from impairing masticatory function, missing teeth can significantly compromise dental bone and surrounding tissues. Dental implantation treatment (Fig. 19c), a leading prosthetic technology, has witnessed rapid development over the past five decades and stands as a primary recourse when the application of dental bridges is precluded by adjacent teeth. A multitude of studies in the literature have leveraged ML techniques to explore dental implants comprehensively [772,773,774,775,776,777,778,779,780,781], as catalogued in Table 14.
The failure of dental implants is subject to a myriad of complex factors, encompassing patient-specific variables, implant positioning, surrounding bony tissue condition, fixture characteristics, surgical procedures, and implant timing. Conventional statistical methods encounter limitations due to the diverse types of data, whereas physics-based models addressing these intricate factors are often impractical. Consequently, researchers have turned to ML-based regression models, constituting a dynamic domain in dental implant research [762, 782, 783]. Notably, fracture emerges as a common failure mode for dental implants. Lee et al. [763] utilised a convolutional neural network (CNN) model to detect and classify fractured dental implants through radiograph images, presenting a potential real-time clinical application.
In order to address another critical issue that peri-implant bone loss leads to implant loosening, Cha et al. [764] employed a CNN model to measure peri-implant bone loss from radiographs, offering an in situ assessment of marginal bone loss severity. Additionally, Zhang et al. [765] identified trabeculae microstructure parameters as effective indicators for marginal bone loss. Comparative studies employing various regression models, including SVM, ANN, logistic regression (LR), and RF, revealed the superior accuracy of SVM in predicting marginal bone loss.
ML approaches have also found applications in the patient-specific design of dental implants. For instance, Hsu et al. [168] utilised the deep learning U-net neural networks and an ANN model for mechanobiological design, considering peri-implant bone healing, remodelling, and cell proliferation to optimise designs and prevent potential failures due to bone loss.
6.4 Arterial Stents
Stent treatment stands as a favoured therapeutic strategy for numerous cardiovascular patients seeking to restore blood vessel circulation. These miniature mesh-like mechanical devices, known as stents (Fig. 19d), compress plaque against vessel walls, opening obstructed arteries and offering substantial benefits by minimising surgical risks and reducing hospitalisation periods. Several studies have investigated the applications of ML techniques in arterial stents, as outlined in Table 14. Typically crafted from biomaterials such as stainless steel, Ni–Ti alloy, cobalt-chromium, titanium, and its alloys, stents are intricately shaped through laser cutting techniques. Maudes et al. [766] developed a ML-based model to establish the highly nonlinear relationship between processing parameters and geometrical characteristics during the manufacturing process. This approach holds promise for reducing the high costs associated with experimental tests.
Stent expansion in arteries is pivotal for ensuring clinical outcomes, where inadequate expansion and malposition can lead to chronic complications such as restenosis and thrombosis. Dong et al. [767] integrated ML models with FE methods to predict stent expansion in a calcified coronary artery. Patient-specific optical coherence tomography (OCT) images were employed to reconstruct the FE models, in which the cross-sectional images of pre- and post-stenting were used as a training dataset. Eight different features extracted from pre-stenting images served as inputs for the support vector regression (SVR) model, predicting lumen area from post-stenting images.
7 Challenges and Perspectives
ML techniques present unprecedented opportunities for exploring novel biomaterials, modelling biomechanics and mechanobiology, and designing and controlling biofabrication processes due to their robust capabilities in handling significantly large and complex datasets. The integration of ML approaches in the biomedical engineering field holds significant promise for the entire framework, encompassing biomaterials design, in silico biomechanical or mechanobiological modelling, and the fabrication of real-life products, thereby facilitating the development of novel patient-specific treatments with minimal side effects for clinical applications. However, several key challenges and perspectives in integrating ML approaches for biomedical engineering issues need to be addressed:
-
(1)
Data collection and preparation: The availability of useful and comprehensive datasets is paramount for training ML models. While training data can be sourced from either experimental tests or computational simulations, experimental tests are often expensive, and simulations for novel materials based on universal physical laws can often be demanding. Moreover, data labelling for supervised learning may incur significant costs. Therefore, the collection and preparation of reliable data without sacrificing crucial information are essential for the effective implementation of ML techniques.
-
(2)
ML-based design approaches: Conventional approaches for biomaterials design often rely on physical/chemical intuition. In contrast, ML-based design approaches can learn from data, unravelling the relationship between material structures and mechanical/biological behaviours. This offers an efficient method enabling brute force design, which is often prohibitive in conventional approaches due to excessive computational costs.
-
(3)
Data-driven modelling for Biomechanics and Mechanobiology: Data-driven and ML-based modelling for biomechanics and mechanobiology create new opportunities for the development of digital twins. These twins can precisely replicate in vitro and/or in vivo behaviours of biological systems in silico, offering a supplementary yet important approach to in vitro and/or in vivo tests or guidelines for practical clinics.
-
(4)
Integration of physics laws in ML models: Embedding physics laws in ML models could enhance the learning process from data, especially in mechanics-related fields. Conversely, ML approaches are expected to discover new physical laws behind data and integrate them into conventional frameworks for more reliable and more realistic solutions.
-
(5)
ML-based approaches in bioprinting: The design for bioprinting is still in its early stages. ML-based approaches illuminate the design optimisation of biomedical devices, potentially overcoming challenges associated with conventional optimisation methods. Additionally, ML-based techniques provide a feasible way to establish a closed-loop fabrication process with minimal human intervention.
-
(6)
Interdisciplinary knowledge in biomedical engineering: Biomedical engineering involves complex multidisciplinary knowledge and methodologies, commonly posing great challenges and high cost for the development and implementation of novel treatments for critical healthcare issues. Advanced ML-based methods are expected to create a comprehensive framework in futuristic studies—from the design of novel biomaterials to modelling biomechanical and mechanobiological behaviours, to the fabrication of real-life products.
8 Conclusion
This paper provides an overview of the state-of-the-art of ML approaches in the biomedical engineering field. It begins with a brief review of various ML approaches, followed by discussions on their applications in biomaterials, biomechanics, mechanobiology, and biofabrication, respectively. The review emphasises their advantages, such as superior data processing and data mining capabilities, development of digital twins, and innovative design of biomaterials. The exploration extends from hard tissue to soft tissue, monoscale to multiscale perspectives. The current status of ML techniques in biofabrication is also reviewed, shedding light on structural and metamaterial design, process optimisation, and in situ monitoring for AM processes. The paper concludes by exemplifying typical biomedical applications using ML-based approaches, broadening the horizon for future work, and addressing challenges and perspectives. This review aims to provide valuable information and lighten key perspectives for researchers, engineers, and clinicians, navigating this rapidly emerging multidisciplinary field of ML in biomedical engineering.
Abbreviations
- ABS:
-
Acrylonitrile butadiene styrene
- AM:
-
Additive manufacturing
- ANN:
-
Artificial neural network
- AR-RF:
-
Additive regression random forest
- BJ:
-
Binder jetting
- BO:
-
Bayesian optimisation
- BPNN:
-
Backpropagation neural network
- CFSFDP:
-
Clustering by fast search and finding of density peaks
- cGAN:
-
Conditional generative adversarial network
- CNN:
-
Convolutional neural networks
- DED:
-
Directed energy deposition
- DIC:
-
Digital image correlation
- DLP:
-
Digital light processing
- DMO:
-
Discrete material optimisation
- DOD:
-
Drop-on-demand
- DOE:
-
Design of experiment
- DT:
-
Decision tree
- EBB:
-
Extrusion-based bioprinting
- EBM:
-
Electron beam melting
- EBSD:
-
Electron backscatter diffraction
- EDS:
-
Energy dispersive X-ray spectroscopy
- EN:
-
Elastic nets
- FDM:
-
Fused deposition modelling
- FE:
-
Finite element
- FE2 :
-
Multiscale finite element
- FFF:
-
Fused filament fabrication
- GA:
-
Genetic algorithm
- GAGP:
-
Globally approximate Gaussian process
- GAN:
-
Generative adversarial network
- GB:
-
Gradient boosting
- GBDT:
-
Gradient boosting regression tree
- GEP:
-
Gene expression programming
- GNN:
-
Graph neural network
- GP:
-
Genetic programming
- GPR:
-
Gaussian process regression
- Grad-CAM:
-
Gradient-weighted class activation mapping
- GRU:
-
Gated recurrent unit network
- HA:
-
Hydroxyapatite
- hEB:
-
Human embryonic stem cell embryoid bodies
- hiMSC:
-
Human immortalised mesenchymal stem cell
- IFT:
-
Invariant feature transform
- IND:
-
Indomethacin
- KEM:
-
Knee extension moment
- KFA:
-
Knee flexion angle
- KNN:
-
K-nearest neighbour learning
- KPA:
-
Knee power absorption
- KRR:
-
Kernel ridge regression
- LM:
-
Levenberg–Marquardt
- LR:
-
Linear regression
- LSR:
-
Lasso regression
- LSTM:
-
Long short-term memory
- LVGP:
-
Latent-variable Gaussian process
- ML:
-
Machine learning
- MLP:
-
Multilayer perceptron
- MMC:
-
Moving morphable component
- MP-LVGP:
-
Multi-response latent-variable Gaussian process
- MS:
-
Mass spectrumetry
- NB:
-
Naïve Bayesian
- NN:
-
Neural networks
- NSAID:
-
Non-steroidal anti-inflammatory drug
- NST:
-
Neural style transfer
- OSS:
-
One-step secant
- PBF:
-
Powder bed fusion
- PCA:
-
Principal component analysis
- PINN:
-
Physics-informed neural network
- PLA:
-
Polylactic acid or polylactide
- PLGA:
-
Poly (lactic-co-glycolic acid)
- RF:
-
Random forest
- RNN:
-
Recurrent neural network
- ROI:
-
Region of interest
- RR:
-
Ridge regression
- RVE:
-
Representative volume element
- S3VM:
-
Semi-supervised support vector machine
- SCG:
-
Scaled conjugate gradient
- SIMP:
-
Solid isotropic material with penalisation
- SLA:
-
Stereolithography
- SLM:
-
Selective laser melting
- SLS:
-
Selective laser sintering
- SOM:
-
Self-organising map
- SVM:
-
Support vector machine
- TB:
-
Trabecular bone
- TNT:
-
Titanium dioxide nanotube
- TPMS:
-
Triply periodic minimal surface
- VAE:
-
Variational autoencoders
- vGRF:
-
Vertical ground reaction force
- WAAM:
-
Wire arc additive manufacturing
- XGBoost:
-
Extreme gradient boosting
References
Campoccia D, Montanaro L, Arciola CR (2013) A review of the biomaterials technologies for infection-resistant surfaces. Biomaterials 34(34):8533–8554
Roach P, Eglin D, Rohde K, Perry CC (2007) Modern biomaterials: a review—bulk properties and implications of surface modifications. J Mater Sci Mater Med 18(7):1263–1277
Hubbell JA (1995) Biomaterials in tissue engineering. Biotechnology 13(6):565–576
Carter DR, Beaupre GS, Giori NJ, Helms JA (1998) Mechanobiology of skeletal regeneration. Clin Orthop Relat Res 355S:S41–S55
Knudson D (2007) Fundamentals of biomechanics and qualitative analysis, fundamental of biomechanics. Springer, pp 23–37
Nordin M, Frankel V (1991) Basic biomechanics of the musculoskeletal systm. J Pediatric Orthopaedics 11(6):788
Tyler WJ (2012) The mechanobiology of brain function. Nat Rev Neurosci 13(12):867–878
Mandrycky C, Wang Z, Kim K, Kim DH (2016) 3D bioprinting for engineering complex tissues. Biotechnol Adv 34(4):422–434
Murphy SV, Atala A (2014) 3D bioprinting of tissues and organs. Nat Biotechnol 32(8):773–785
Franz S, Rammelt S, Scharnweber D, Simon JC (2011) Immune responses to implants—a review of the implications for the design of immunomodulatory biomaterials. Biomaterials 32(28):6692–6709
Kaur M, Singh K (2019) Review on titanium and titanium based alloys as biomaterials for orthopaedic applications. Mater Sci Eng C Mater Biol Appl 102:844–862
Hollister SJ (2005) Porous scaffold design for tissue engineering. Nat Mater 4(7):518–524
Seneviratne S, Hu YN, Nguyen T, Lan GH, Khalifa S, Thilakarathna K, Hassan M, Seneviratne A (2017) A survey of wearable devices and challenges. IEEE Commun Surv Tutor 19(4):2573–2620
Petit-Zeman S (2001) Regenerative medicine. Nat Biotechnol 19(3):201–206
Langer R (1990) New methods of drug delivery. Science 249(4976):1527–1533
Kohn J (2004) New approaches to biomaterials design. Nat Mater 3(11):745–747
Kohn J, Welsh WJ, Knight D (2007) A new approach to the rationale discovery of polymeric biomaterials. Biomaterials 28(29):4171–4177
Oftadeh R, Perez-Viloria M, Villa-Camacho JC, Vaziri A, Nazarian A (2015) Biomechanics and mechanobiology of trabecular bone: a review. J Biomech Eng 137(1):0108021–01080215
Sherifova S, Holzapfel GA (2019) Biomechanics of aortic wall failure with a focus on dissection and aneurysm: a review. Acta Biomater 99:1–17
Wang C, Li S, Ademiloye AS, Nithiarasu P (2021) Biomechanics of cells and subcellular components: a comprehensive review of computational models and applications. Int J Numer Method Biomed Eng 37(12):e3520
Bielajew BJ, Hu JC, Athanasiou KA (2020) Collagen: quantification, biomechanics, and role of minor subtypes in cartilage. Nat Rev Mater 5(10):730–747
Sahin S, Cehreli MC, Yalcin E (2002) The influence of functional forces on the biomechanics of implant-supported prostheses—a review. J Dent 30(7–8):271–282
Al-Anouti F, Taha Z, Shamim S, Khalaf K, Al-Kaabi L, Alsafar H (2019) An insight into the paradigms of osteoporosis: from genetics to biomechanics. Bone Rep 11:100216
Urbanczyk M, Layland SL, Schenke-Layland K (2020) The role of extracellular matrix in biomechanics and its impact on bioengineering of cells and 3D tissues. Matrix Biol 85–86:1–14
Garcia-Aznar JM, Nasello G, Hervas-Raluy S, Perez MA, Gomez-Benito MJ (2021) Multiscale modeling of bone tissue mechanobiology. Bone 151:116032
Guilak F, Butler DL, Goldstein SA, Baaijens FP (2014) Biomechanics and mechanobiology in functional tissue engineering. J Biomech 47(9):1933–1940
Mullender M, El Haj AJ, Yang Y, van Duin MA, Burger EH, Klein-Nulend J (2004) Mechanotransduction of bone cells in vitro: mechanobiology of bone tissue. Med Biol Eng Comput 42(1):14–21
Wang JH, Thampatty BP (2006) An introductory review of cell mechanobiology. Biomech Model Mechanobiol 5(1):1–16
Zhang X, Kim TH, Thauland TJ, Li H, Majedi FS, Ly C, Gu Z, Butte MJ, Rowat AC, Li S (2020) Unraveling the mechanobiology of immune cells. Curr Opin Biotechnol 66:236–245
Humphrey JD (2003) Review paper: continuum biomechanics of soft biological tissues. Proc R Soc Lond Ser A 459(2029):3–46
Adams MA, Dolan P (2005) Spine biomechanics. J Biomech 38(10):1972–1983
Harper CE, Hernandez CJ (2020) Cell biomechanics and mechanobiology in bacteria: challenges and opportunities. APL Bioeng 4(2):021501
Wu C, Entezari A, Zheng K, Fang J, Zreiqat H, Steven GP, Swain MV, Li Q (2021) A machine learning-based multiscale model to predict bone formation in scaffolds. Nat Comput Sci 1(8):532–541
Wu C, Fang J, Entezari A, Sun G, Swain MV, Xu Y, Steven GP, Li Q (2021) A time-dependent mechanobiology-based topology optimization to enhance bone growth in tissue scaffolds. J Biomech 117:110233
Laz PJ, Browne M (2010) A review of probabilistic analysis in orthopaedic biomechanics. Proc Inst Mech Eng H 224(8):927–943
Saxby DJ, Killen BA, Pizzolato C, Carty CP, Diamond LE, Modenese L, Fernandez J, Davico G, Barzan M, Lenton G, da Luz SB, Suwarganda E, Devaprakash D, Korhonen RK, Alderson JA, Besier TF, Barrett RS, Lloyd DG (2020) Machine learning methods to support personalized neuromusculoskeletal modelling. Biomech Model Mechanobiol 19(4):1169–1185
Lin D, Li Q, Li W, Swain M (2009) Dental implant induced bone remodeling and associated algorithms. J Mech Behav Biomed Mater 2(5):410–432
Wu C, Zheng KK, Fang JG, Steven GP, Li Q (2020) Time-dependent topology optimization of bone plates considering bone remodeling. Comput Methods Appl Mech Eng 359:112702
Wan B, Entezari A, Zhang Z, Wilson T, Yoda N, Zheng K, Wu C, Sun G, Sasaki K, Swain M, Li Q (2021) On fatigue failure prediction of prosthetic devices through XFEM analysis. Int J Fatigue 147:106160
Tammareddi S, Li Q (2010) Effects of material on the deployment of coronary stents. Multi-Funct Mater Struct Iii, Pts 1 and 2 123–125: 315–318.
Tammareddi S, Sun GY, Li Q (2016) Multiobjective robust optimization of coronary stents. Mater Des 90:682–692
Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A (2016) Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 110:45–59
Moroni L, Boland T, Burdick JA, De Maria C, Derby B, Forgacs G, Groll J, Li Q, Malda J, Mironov VA, Mota C, Nakamura M, Shu W, Takeuchi S, Woodfield TBF, Xu T, Yoo JJ, Vozzi G (2018) Biofabrication: a guide to technology and terminology. Trends Biotechnol 36(4):384–402
Bose S, Vahabzadeh S, Bandyopadhyay A (2013) Bone tissue engineering using 3D printing. Mater Today 16(12):496–504
Tappa K, Jammalamadaka U, Weisman JA, Ballard DH, Wolford DD, Pascual-Garrido C, Wolford LM, Woodard PK, Mills DK (2019) 3D printing custom bioactive and absorbable surgical screws, pins, and bone plates for localized drug delivery. J Funct Biomater 10(2):17
Yang F, Chen C, Zhou Q, Gong Y, Li R, Li C, Klampfl F, Freund S, Wu X, Sun Y, Li X, Schmidt M, Ma D, Yu Y (2017) Laser beam melting 3D printing of Ti6Al4V based porous structured dental implants: fabrication, biocompatibility analysis and photoelastic study. Sci Rep 7:45360
Kalkal A, Kumar S, Kumar P, Pradhan R, Willander M, Packirisamy G, Kumar S, Malhotra BD (2021) Recent advances in 3D printing technologies for wearable (bio)sensors. Addit Manuf 46:102088
Bikas H, Stavropoulos P, Chryssolouris G (2015) Additive manufacturing methods and modelling approaches: a critical review. Int J Adv Manuf Technol 83(1–4):389–405
Frazier WE (2014) Metal additive manufacturing: a review. J Mater Eng Perform 23(6):1917–1928
Castro e Costa E, Duarte JP, Bártolo P (2017) A review of additive manufacturing for ceramic production. Rapid Prototyp J 23(5):954–963
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Zhou T, Song Z, Sundmacher K (2019) Big data creates new opportunities for materials research: a review on methods and applications of machine learning for materials design. Engineering 5(6):1017–1026
Johnson NS, Vulimiri PS, To AC, Zhang X, Brice CA, Kappes BB, Stebner AP (2020) Invited review: machine learning for materials developments in metals additive manufacturing. Addit Manuf 36:101641
Qu K, Guo F, Liu X, Lin Y, Zou Q (2019) Application of machine learning in microbiology. Front Microbiol 10:827
Bzdok D, Krzywinski M, Altman N (2018) Machine learning: supervised methods. Nat Methods 15(1):5–6
Celebi ME, Aydin K (2016) Unsupervised learning algorithms. Springer
Belkin M, Niyogi P (2004) Semi-supervised learning on Riemannian manifolds. Mach Learn 56(1–3):209–239
Glorennec PY (2000) Reinforcement learning: an overview, proceedings european symposium on intelligent techniques (ESIT-00). Aachen, Germany, Citeseer, pp 14–15
Fix E, Hodges JL (1951) Discriminatory analysis: nonparametric discrimination: consistency properties. PsycEXTRA Dataset, American Psychological Association (APA)
Cover TM, Hart PE (1967) Nearest neighbor pattern classification. IEEE Trans Inf Theory 13(1):21
Won Yoon J, Friel N (2015) Efficient model selection for probabilistic K nearest neighbour classification. Neurocomputing 149:1098–1108
Surya V, Haneen P, Ahmad A, Omar B, Ahmad L (2019) Effects of distance measure choice on KNN classifier performance—a review. Mary Ann Liebert
Schwenker F, Kestler HA, Palm G (2001) Three learning phases for radial-basis-function networks. Neural Netw 14(4–5):439–458
Kang SK (2021) Nearest neighbor learning with graph neural networks. Mathematics 9(8):830
Kataria A, Singh M (2013) A review of data classification using k-nearest neighbour algorithm. Int J Emerg Technol Adv Eng 3(6):354–360
Bodduluri S, Newell JD Jr, Hoffman EA, Reinhardt JM (2013) Registration-based lung mechanical analysis of chronic obstructive pulmonary disease (COPD) using a supervised machine learning framework. Acad Radiol 20(5):527–536
Sarker IH, Faruque F, Alqahtani H, Kalim A (2018) K-nearest neighbor learning based diabetes mellitus prediction and analysis for eHealth services. EAI Endorsed Trans Scalable Inf Syst 7(26)
Chandel K, Kunwar V, Sabitha S, Choudhury T, Mukherjee S (2017) A comparative study on thyroid disease detection using K-nearest neighbor and Naive Bayes classification techniques. CSI Trans ICT 4(2–4):313–319
Salzberg SL (1994) C4.5: programs for machine learning by J. Ross Quinlan: Morgan Kaufmann Publishers, Inc, 1993. Mach Learn 16(3):235–240
Quinlan JR (1999) Simplifying decision trees. Int J Hum Comput Stud 51(2):497–510
Quinlan JR (1986) Induction of decision trees. Mach Learn 1(1):81–106
Connaboy C, Eagle SR, Johnson CD, Flanagan SD, Mi QI, Nindl BC (2019) Using machine learning to predict lower-extremity injury in US special forces. Med Sci Sports Exerc 51(5):1073–1079
Liu Q, Mo S, Cheung VCK, Cheung BMF, Wang S, Chan PPK, Malhotra A, Cheung RTH, Chan RHM (2020) Classification of runners’ performance levels with concurrent prediction of biomechanical parameters using data from inertial measurement units. J Biomech 112:110072
Martinez-Martinez F, Ruperez-Moreno MJ, Martinez-Sober M, Solves-Llorens JA, Lorente D, Serrano-Lopez AJ, Martinez-Sanchis S, Monserrat C, Martin-Guerrero JD (2017) A finite element-based machine learning approach for modeling the mechanical behavior of the breast tissues under compression in real-time. Comput Biol Med 90:116–124
Matijevich ES, Volgyesi P, Zelik KE (2021) A promising wearable solution for the practical and accurate monitoring of low back loading in manual material handling. Sensors 21(2):340
Moufawad el Achkar C, Lenoble-Hoskovec C, Paraschiv-Ionescu A, Major K, Bula C, Aminian K (2016) Instrumented shoes for activity classification in the elderly. Gait Posture 44:12–17
Nathan R, Spiegel O, Fortmann-Roe S, Harel R, Wikelski M, Getz WM (2012) Using tri-axial acceleration data to identify behavioral modes of free-ranging animals: general concepts and tools illustrated for griffon vultures. J Exp Biol 215(Pt 6):986–996
Whiteside D, Reid M (2017) Spatial characteristics of professional tennis serves with implications for serving aces: a machine learning approach. J Sports Sci 35(7):648–654
Breiman L (1996) Bagging predictors. Mach Learn 24(2):123–140
Biau G, Scornet E (2016) A random forest guided tour. TEST 25(2):197–227
Ahamed NU, Kobsar D, Benson L, Clermont C, Kohrs R, Osis ST, Ferber R (2018) Using wearable sensors to classify subject-specific running biomechanical gait patterns based on changes in environmental weather conditions. PLoS ONE 13(9):e0203839
Chalitsios C, Nikodelis T, Konstantakos V, Kollias I (2022) Sensitivity of movement features to fatigue during an exhaustive treadmill run. Eur J Sport Sci 22(9):1374–1382
Hua A, Chaudhari P, Johnson N, Quinton J, Schatz B, Buchner D, Hernandez ME (2020) Evaluation of machine learning models for classifying upper extremity exercises using inertial measurement unit-based kinematic data. IEEE J Biomed Health Inform 24(9):2452–2460
Wahid F, Begg RK, Hass CJ, Halgamuge S, Ackland DC (2015) Classification of Parkinson’s disease gait using spatial-temporal gait features. IEEE J Biomed Health Inform 19(6):1794–1802
Flach PA, Lachiche N (2004) Naive Bayesian classification of structured data. Mach Learn 57(3):233–269
Chen SL, Webb GI, Liu LY, Ma X (2020) A novel selective Naive Bayes algorithm. Knowl-Based Syst 192:105361
Singh BK (2019) Determining relevant biomarkers for prediction of breast cancer using anthropometric and clinical features: a comparative investigation in machine learning paradigm. Biocybern Biomed Eng 39(2):393–409
Svensson CM, Krusekopf S, Lucke J, Thilo Figge M (2014) Automated detection of circulating tumor cells with naive Bayesian classifiers. Cytometry A 85(6):501–511
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24(12):1565–1567
Huang G, Huang GB, Song S, You K (2015) Trends in extreme learning machines: a review. Neural Netw 61:32–48
Fatima S, Srinivasu B (2017) Text document categorization using support vector machine. Int Res J Eng Technol 4(2):141–147
Okwuashi O, Ndehedehe CE (2020) Deep support vector machine for hyperspectral image classification. Pattern Recogn 103:107298
Gokulnath CB, Shantharajah SP (2018) An optimized feature selection based on genetic approach and support vector machine for heart disease. Clust Comput 22(S6):14777–14787
Cust EE, Sweeting AJ, Ball K, Robertson S (2019) Machine and deep learning for sport-specific movement recognition: a systematic review of model development and performance. J Sports Sci 37(5):568–600
Phinyomark A, Petri G, Ibanez-Marcelo E, Osis ST, Ferber R (2018) Analysis of big data in gait biomechanics: current trends and future directions. J Med Biol Eng 38(2):244–260
Baghdadi A, Megahed FM, Esfahani ET, Cavuoto LA (2018) A machine learning approach to detect changes in gait parameters following a fatiguing occupational task. Ergonomics 61(8):1116–1129
Chakravarty P, Cozzi G, Ozgul A, Aminian K (2019) A novel biomechanical approach for animal behaviour recognition using accelerometers. Methods Ecol Evol 10(6):802–814
Clermont CA, Osis ST, Phinyomark A, Ferber R (2017) Kinematic gait patterns in competitive and recreational runners. J Appl Biomech 33(4):268–276
Haudenschild AK, Sherlock BE, Zhou X, Hu JC, Leach JK, Marcu L, Athanasiou KA (2019) Non-destructive detection of matrix stabilization correlates with enhanced mechanical properties of self-assembled articular cartilage. J Tissue Eng Regen Med 13(4):637–648
Zahra SB, Khan MA, Abbas S, Khan KM, Al-Ghamdi MA, Almotiri SH (2021) Marker-based and marker-less motion capturing video data: person and activity identification comparison based on machine learning approaches. Comput Mater Continua 66(2):1269–1282
Dreiseitl S, Ohno-Machado L (2002) Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform 35(5–6):352–359
Maulud D, Abdulazeez AM (2020) A review on linear regression comprehensive in machine learning. J Appl Sci Technol Trends 1(4):140–147
Shipe ME, Deppen SA, Farjah F, Grogan EL (2019) Developing prediction models for clinical use using logistic regression: an overview. J Thorac Dis 11(Suppl 4):S574–S584
Miguel-Hurtado O, Stevenage S, Bevan C, Guest R (2016) Predicting sex as a soft-biometrics from device interaction swipe gestures. Pattern Recogn Lett 79:44–51
Kim S, Ku S, Chang W, Song JW (2020) Predicting the direction of US stock prices using effective transfer entropy and machine learning techniques. IEEE Access 8:111660–111682
Learning M (2017) Heart disease diagnosis and prediction using machine learning and data mining techniques: a review. Adv Comput Sci Technol 10(7):2137–2159
Agatonovic-Kustrin S, Beresford R (2000) Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J Pharm Biomed Anal 22(5):717–727
Hagan MT, Menhaj MB (1994) Training feedforward networks with the Marquardt algorithm. IEEE Trans Neural Netw 5(6):989–993
Møller MF (1993) A scaled conjugate gradient algorithm for fast supervised learning. Neural Netw 6(4):525–533
Battiti R (1992) First- and second-order methods for learning: between steepest descent and newton’s method. Neural Comput 4(2):141–166
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adam M, Gertych A, Tan RS (2017) A deep convolutional neural network model to classify heartbeats. Comput Biol Med 89:389–396
Becker S, Plumbley M (1996) Unsupervised neural network learning procedures for feature extraction and classification. Appl Intell 6(3):185–203
Cannistraci CV, Ravasi T, Montevecchi FM, Ideker T, Alessio M (2010) Nonlinear dimension reduction and clustering by minimum curvilinearity unfold neuropathic pain and tissue embryological classes. Bioinformatics 26(18):i531–i539
Bradley PS, Fayyad UM (1998) Refining initial points for k-means clustering. ICML, Citeseer, pp 91–99
Kohonen T (1998) The self-organizing map. Neurocomputing 21(1–3):1–6
Ringnér M (2008) What is principal component analysis? Nat Biotechnol 26(3):303–304
Jati A, Georgiou P (2019) Neural predictive coding using convolutional neural networks toward unsupervised learning of speaker characteristics. IEEE–-ACM Trans Audio Speech Lang Process 27(10):1577–1589
MacQueen J (1967) Some methods for classification and analysis of multivariate observations. In: Proceedings of the fifth Berkeley symposium on mathematical statistics and probability, Oakland, pp. 281–297
Kohonen T, Somervuo P (1998) Self-organizing maps of symbol strings. Neurocomputing 21(1–3):19–30
Kohonen T (2013) Essentials of the self-organizing map. Neural Netw 37:52–65
Aljohani M, Kipp K (2020) Use of self-organizing maps to study sex- and speed-dependent changes in running biomechanics. Hum Mov Sci 72:102649
Nilashi M, Ibrahim O, Ahmadi H, Shahmoradi L, Farahmand M (2018) A hybrid intelligent system for the prediction of Parkinson’s disease progression using machine learning techniques. Biocybern Biomed Eng 38(1):1–15
Liu BC, Lai MZ, Wu JL, Fu CC, Binaykia A (2020) Patent analysis and classification prediction of biomedicine industry: SOM-KPCA-SVM model. Multimedia Tools App 79(15–16):10177–10197
Hasan S, Shamsuddin SM (2019) Multi-strategy learning and deep harmony memory improvisation for self-organizing neurons. Soft Comput 23(1):285–303
Pearson, LIII K (2010) On lines and planes of closest fit to systems of points in space. Lond, Edinburgh, Dublin Philos Magaz J Sci 2(11):559–572
Lawrence N (2005) Probabilistic non-linear principal component analysis with Gaussian process latent variable models. J Mach Learn Res 6(11):1783–1816
Wold S, Esbensen K, Geladi P (1987) Principal component analysis: chimometrics and intelligent laboratory systems. In: IEEE conference on emerging technologies & factory automation. Efta, pp. 704–706
Gokgoz E, Subasi A (2015) Comparison of decision tree algorithms for EMG signal classification using DWT. Biomed Signal Process Control 18:138–144
Phinyomark A, Osis ST, Hettinga BA, Kobsar D, Ferber R (2016) Gender differences in gait kinematics for patients with knee osteoarthritis. BMC Musculoskelet Disord 17(1):157
Dong GG, Liao GS, Liu HW, Kuang GY (2018) A review of the autoencoder and its variants. IEEE Geosci Remote Sens Magaz 6(3):44–68
Mnih V, Larochelle H, Hinton GE (2012) Conditional restricted Boltzmann machines for structured output prediction. arXiv preprint arXiv:1202.3748
Li J, Jia J, Xu D (2018) Unsupervised representation learning of image-based plant disease with deep convolutional generative adversarial networks. 2018 37th Chinese Control Conference (CCC). IEEE, pp 9159–9163
Gibson BR, Rogers TT, Zhu X (2013) Human semi-supervised learning. Top Cogn Sci 5(1):132–172
Fooladgar F, Nguyen Nhatto M, Javadi G, Sojoudi S, Eshumani W, Chang S, Black P, Mousavi P, Abolmaesumi P (2023) Semi-supervised learning from coarse histopathology labels. Comput Methods Biomech Biomed Eng 11(4):1143–1150
Teevno MA, Ochoa-Ruiz G, Ali S (2023) A semi-supervised Teacher–Student framework for surgical tool detection and localization. Comput Methods Biomech Biomed Eng 11(4):1033–1041
Schwenker F, Trentin E (2014) Pattern classification and clustering: A review of partially supervised learning approaches. Pattern Recogn Lett 37:4–14
Gan HT, Sang N, Huang R, Tong XJ, Dan ZP (2013) Using clustering analysis to improve semi-supervised classification. Neurocomputing 101:290–298
Piroonsup N, Sinthupinyo S (2018) Analysis of training data using clustering to improve semi-supervised self-training. Knowl-Based Syst 143:65–80
Triguero I, Sáez JA, Luengo J, García S, Herrera F (2014) On the characterization of noise filters for self-training semi-supervised in nearest neighbor classification. Neurocomputing 132:30–41
Li Q, Han Z, Wu X-M (2018) Deeper insights into graph convolutional networks for semi-supervised learning. Thirty-Second AAAI conference on artificial intelligence
Tena A, Claria F, Solsona F, Povedano M (2023) Voiceprint and machine learning models for early detection of bulbar dysfunction in ALS. Comput Methods Programs Biomed 229:107309
Li Y-F, Zhou Z-H (2014) Towards making unlabeled data never hurt. IEEE Trans Pattern Anal Mach Intell 37(1):175–188
Ding S, Zhu Z, Zhang X (2015) An overview on semi-supervised support vector machine. Neural Comput Appl 28(5):969–978
Blum A, Chawla S (2001) Learning from labeled and unlabeled data using graph mincuts
Joachims T (2003) Transductive learning via spectral graph partitioning. In: Proceedings of the 20th international conference on machine learning (ICML-03), pp. 290–297
Zhu X, Ghahramani Z, Lafferty JD, Semi-supervised learning using Gaussian fields and harmonic functions. In: Proceedings of the 20th International conference on Machine learning (ICML-03), pp. 912–919
Karasuyama M, Mamitsuka H (2017) Adaptive edge weighting for graph-based learning algorithms. Mach Learn 106(2):307–335
Neftci EO, Averbeck BB (2019) Reinforcement learning in artificial and biological systems. Nat Mach Intell 1(3):133–143
Lei C (2021) Deep reinforcement learning. Deep learning and practice with MindSpore. Springer, pp 217–243
He B, Zhu XR, Zhang D (2020) Boundary encryption-based Monte Carlo learning method for workspace modeling. J Comput Inf Sci Eng. https://doi.org/10.1115/1.4046816
Jia J, Wang W (2020) Review of reinforcement learning research. 2020 35th Youth Academic Annual Conference of Chinese Association of Automation (YAC). IEEE, pp 186–191
Yassine AA, Lilge L, Betz V (2021) Optimizing interstitial photodynamic therapy planning with reinforcement learning-based diffuser placement. IEEE Trans Biomed Eng 68(5):1668–1679
Banik S, Loefller T, Manna S, Chan HY, Srinivasan S, Darancet P, Hexemer A, Sankaranarayanan SKRS (2023) A Continuous Action Space Tree search for INverse desiGn (CASTING) framework for materials discovery. NPJ Comput Mater 9(1):177
Liu J (2021) On the convergence of reinforcement learning with Monte Carlo exploring starts. Automatica 129:109693
Watkins CJCH, Dayan P (1992) Q-learning. Mach Learn 8(3–4):279–292
Barrett CD, Suzuki Y, Hussein S, Garg L, Tumolo A, Sandhu A, West JJ, Zipse M, Aleong R, Varosy P, Tzou WS, Banaei-Kashani F, Rosenberg MA (2023) Evaluation of quantitative decision-making for rhythm management of atrial fibrillation using tabular Q-learning. J Am Heart Assoc 12(9):e028483
Zhou CM, Huang BD, Fränti P (2022) A review of motion planning algorithms for intelligent robots. J Intell Manuf 33(2):387–424
Zhang JJ, Zhang C, Chien WC (2021) Overview of deep reinforcement learning improvements and applications. J Internet Technol 22(2):239–255
Dee KC, Puleo DA, Bizios R (2002) An introduction to tissue-biomaterial interactions. John Wiley & Sons Inc
Przekora A (2019) The summary of the most important cell-biomaterial interactions that need to be considered during in vitro biocompatibility testing of bone scaffolds for tissue engineering applications. Mater Sci Eng C Mater Biol Appl 97:1036–1051
Yadav P, Yadav H, Shah VG, Shah G, Dhaka G (2015) Biomedical biopolymers, their origin and evolution in biomedical sciences: a systematic review. J Clin Diagn Res 9(9):21–25
Mtibe A, Motloung MP, Bandyopadhyay J, Ray SS (2021) Synthetic biopolymers and their composites: advantages and limitations-an overview. Macromol Rapid Commun 42(15):e2100130
Miguez-Pacheco V, Hench LL, Boccaccini AR (2015) Bioactive glasses beyond bone and teeth: emerging applications in contact with soft tissues. Acta Biomater 13:1–15
Jagur-Grodzinski J (2006) Polymers for tissue engineering, medical devices, and regenerative medicine: concise general review of recent studied. Polym Adv Technol 17(6):395–418
Prasad K, Bazaka O, Chua M, Rochford M, Fedrick L, Spoor J, Symes R, Tieppo M, Collins C, Cao A, Markwell D, Ostrikov KK, Bazaka K (2017) Metallic biomaterials: current challenges and opportunities. Materials 10(8):884
da Mata Santos RP, Vieira Oliveira Prado HE, Soares Aranha Neto I, de Oliveira GAA, Vespasiano Silva AI, Zenobio EG, Manzi FR (2021) Automated identification of dental implants using artificial intelligence. Int J Oral Maxillofac Implants 36(5):918–923
Ghensi P, Manghi P, Zolfo M, Armanini F, Pasolli E, Bolzan M, Bertelle A, Dell’Acqua F, Dellasega E, Waldner R, Tessarolo F, Tomasi C, Segata N (2020) Strong oral plaque microbiome signatures for dental implant diseases identified by strain-resolution metagenomics. NPJ Biofilms Microbiomes 6(1):47
Hsu CW, Yang AC, Kung PC, Tsou NT, Chen NY (2021) Engineer design process assisted by explainable deep learning network. Sci Rep 11(1):22525
Kumar A, Sharma R, Gupta AK (2021) Experimental investigation of WEDM process through integrated desirability and machine learning technique on implant material. J Mech Behav Mater 30(1):38–48
Lee JH, Jeong SN (2020) Efficacy of deep convolutional neural network algorithm for the identification and classification of dental implant systems, using panoramic and periapical radiographs: a pilot study. Medicine 99(26):e20787
Lee JH, Kim YT, Lee JB, Jeong SN (2020) A performance comparison between automated deep learning and dental professionals in classification of dental implant systems from dental imaging: a multi-center study. Diagnostics 10(11):910
Mahri M, Shen N, Berrizbeitia F, Rodan R, Daer A, Faigan M, Taqi D, Wu KY, Ahmadi M, Ducret M, Emami E, Tamimi F (2021) Osseointegration pharmacology: a systematic mapping using artificial intelligence. Acta Biomater 119:284–302
Vyas N, Sammons RL, Addison O, Dehghani H, Walmsley AD (2016) A quantitative method to measure biofilm removal efficiency from complex biomaterial surfaces using SEM and image analysis. Sci Rep 6:32694
Klawitter JJ, Weinstein AM, Cooke FW, Peterson LJ, Pennel BM, McKinney RV Jr (1977) An evaluation of porous alumina ceramic dental implants. J Dent Res 56(7):768–776
Moutos FT, Freed LE, Guilak F (2007) A biomimetic three-dimensional woven composite scaffold for functional tissue engineering of cartilage. Nat Mater 6(2):162–167
Sharma U, Concagh D, Core L, Kuang Y, You C, Pham Q, Zugates G, Busold R, Webber S, Merlo J, Langer R, Whitesides GM, Palasis M (2018) The development of bioresorbable composite polymeric implants with high mechanical strength. Nat Mater 17(1):96–103
Madiona RMT, Winkler DA, Muir BW, Pigram PJ (2019) Optimal machine learning models for robust materials classification using ToF-SIMS data. Appl Surf Sci 487:773–783
Wei Q, Melko RG, Chen JZY (2017) Identifying polymer states by machine learning. Phys Rev E 95(3–1):032504
Burroughs L, Amer MH, Vassey M, Koch B, Figueredo GP, Mukonoweshuro B, Mikulskis P, Vasilevich A, Vermeulen S, Dryden IL, Winkler DA, Ghaemmaghami AM, Rose F, de Boer J, Alexander MR (2021) Discovery of synergistic material-topography combinations to achieve immunomodulatory osteoinductive biomaterials using a novel in vitro screening method: the ChemoTopoChip. Biomaterials 271:120740
Epa VC, Yang J, Mei Y, Hook AL, Langer R, Anderson DG, Davies MC, Alexander MR, Winkler DA (2012) Modelling human embryoid body cell adhesion to a combinatorial library of polymer surfaces. J Mater Chem 22(39):20902–20906
Robles-Bykbaev Y, Naya S, Diaz-Prado S, Calle-Lopez D, Robles-Bykbaev V, Garzon L, Sanjurjo-Rodriguez C, Tarrio-Saavedra J (2019) An artificial-vision- and statistical-learning-based method for studying the biodegradation of type I collagen scaffolds in bone regeneration systems. PeerJ 7:e7233
Rostam HM, Fisher LE, Hook AL, Burroughs L, Luckett JC, Figueredo GP, Mbadugha C, Teo ACK, Latif A, Kämmerling L, Day M, Lawler K, Barrett D, Elsheikh S, Ilyas M, Winkler DA, Alexander MR, Ghaemmaghami AM (2020) Immune-instructive polymers control macrophage phenotype and modulate the foreign body response. Matter 2(6):1564–1581
Santana R, Zuluaga R, Ganan P, Arrasate S, Onieva E, Gonzalez-Diaz H (2020) Predicting coated-nanoparticle drug release systems with perturbation-theory machine learning (PTML) models. Nanoscale 12(25):13471–13483
Vassey MJ, Figueredo GP, Scurr DJ, Vasilevich AS, Vermeulen S, Carlier A, Luckett J, Beijer NRM, Williams P, Winkler DA, de Boer J, Ghaemmaghami AM, Alexander MR (2020) Immune modulation by design: using topography to control human monocyte attachment and macrophage differentiation. Adv Sci 7(11):1903392
Wu K, Sukumar N, Lanzillo NA, Wang C, Ramprasad RR, Ma R, Baldwin AF, Sotzing G, Breneman C (2016) Prediction of polymer properties using infinite chain descriptors (ICD) and machine learning: toward optimized dielectric polymeric materials. J Polym Sci Part B—Polym Phys 54(20):2082–2091
Damiati SA, Damiati S (2021) microfluidic synthesis of indomethacin-loaded PLGA microparticles optimized by machine learning. Front Mol Biosci 8:677547
Chittam S, Gokaraju B, Xu ZG, Sankar J, Roy K (2021) Big data mining and classification of intelligent material science data using machine learning. Appl Sci 11(18):8596
Li MH, Mesbah M, Fallahpour A, Nasiri-Tabrizi B, Liu BY (2021) Mechanical strength estimation of ultrafine-grained magnesium implant by neural-based predictive machine learning. Mater Lett 305:130627
Shen Z, Wang S, Shen Z, Tang Y, Xu J, Lin C, Chen X, Huang Q (2021) Deciphering controversial results of cell proliferation on TiO(2) nanotubes using machine learning. Regen Biomater 8(4):rbab025
He Y, Cubuk ED, Allendorf MD, Reed EJ (2018) Metallic metal-organic frameworks predicted by the combination of machine learning methods and ab initio calculations. J Phys Chem Lett 9(16):4562–4569
Janet JP, Kulik HJ (2017) Resolving transition metal chemical space: feature selection for machine learning and structure-property relationships. J Phys Chem A 121(46):8939–8954
Moghadam PZ, Rogge SMJ, Li A, Chow CM, Wieme J, Moharrami N, Aragones-Anglada M, Conduit G, Gomez-Gualdron DA, Van Speybroeck V, Fairen-Jimenez D (2019) Structure-mechanical stability relations of metal-organic frameworks via machine learning. Matter 1(1):219–234
Toyao T, Suzuki K, Kikuchi S, Takakusagi S, Shimizu K, Takigawa I (2018) Toward effective utilization of methane: machine learning prediction of adsorption energies on metal alloys. J Phys Chem C 122(15):8315–8326
Xiong J, Shi S-Q, Zhang T-Y (2020) A machine-learning approach to predicting and understanding the properties of amorphous metallic alloys. Mater Des 187:108378
Zhang Y, Wen C, Wang CX, Antonov S, Xue DZ, Bai Y, Su YJ (2020) Phase prediction in high entropy alloys with a rational selection of materials descriptors and machine learning models. Acta Mater 185:528–539
Wen C, Zhang Y, Wang CX, Xue DZ, Bai Y, Antonov S, Dai LH, Lookman T, Su YJ (2019) Machine learning assisted design of high entropy alloys with desired property. Acta Mater 170:109–117
Wu CT, Chang HT, Wu CY, Chen SW, Huang SY, Huang MX, Pan YT, Bradbury P, Chou J, Yen HW (2020) Machine learning recommends affordable new Ti alloy with bone-like modulus. Mater Today 34:41–50
Tripathi G, Anowarul H, Agarwal K, Prasad DK (2019) Classification of micro-damage in piezoelectric ceramics using machine learning of ultrasound signals. Sensors 19(19):4216
Vallejos Baier R, Benjumeda Wijnhoven I, Del Valle VI, Millan Giovanetti C, Vivanco JF (2019) Microporosity clustering assessment in calcium phosphate bioceramic particles. Front Bioeng Biotechnol 7:281
Gopinath KGS, Pal S, Tambe P (2018) Prediction of weight percentage alumina and pore volume fraction in bio-ceramics using Gaussian process regression and minimax probability machine regression. Mater Today—Proc 5(5):12233–12239
Qin JC, Liu ZF, Ma MS, Li YX (2021) Machine learning approaches for permittivity prediction and rational design of microwave dielectric ceramics. J Materiom 7(6):1284–1293
Qu N, Liu Y, Liao MQ, Lai ZH, Zhou F, Cui PC, Han TY, Yang DN, Zhu JC (2019) Ultra-high temperature ceramics melting temperature prediction via machine learning. Ceram Int 45(15):18551–18555
Yang P, Wu SS, Wu HN, Lu DL, Zou WJ, Chu LJ, Shao YZ, Wu SH (2021) Prediction of bending strength of SiN4 using machine learning. Ceram Int 47(17):23919–23926
Kaufmann K, Maryanovsky D, Mellor WM, Zhu CY, Rosengarten AS, Harrington TJ, Oses C, Toher C, Curtarolo S, Vecchio KS (2020) Discovery of high-entropy ceramics via machine learning. NPJ Comput Mater. https://doi.org/10.1038/s41524-020-0317-6
Yuan R, Tian Y, Xue D, Xue D, Zhou Y, Ding X, Sun J, Lookman T (2019) Accelerated search for BaTiO(3)-based ceramics with large energy storage at low fields using machine learning and experimental design. Adv Sci 6(21):1901395
AmirSiddiq M (2020) Data-driven finite element method: theory and applications. Proc Inst Mech Eng Part C: J Mech Eng Sci 235(17):3329–3339
Yang ZJ, Yabansu YC, Al-Bahrani R, Liao WK, Choudhary AN, Kalidindi SR, Agrawal A (2018) Deep learning approaches for mining structure-property linkages in high contrast composites from simulation datasets. Comput Mater Sci 151:278–287
Chen CT, Gu GX (2019) Effect of constituent materials on composite performance: exploring design strategies via machine learning. Adv Theory Simulations 2(6):1900056
Gu GX, Chen CT, Richmond DJ, Buehler MJ (2018) Bioinspired hierarchical composite design using machine learning: simulation, additive manufacturing, and experiment. Mater Horiz 5(5):939–945
Han T, Stone-Weiss N, Huang J, Goel A, Kumar A (2020) Machine learning as a tool to design glasses with controlled dissolution for healthcare applications. Acta Biomater 107:286–298
Yu CH, Qin Z, Buehler MJ (2019) Artificial intelligence design algorithm for nanocomposites optimized for shear crack resistance. Nano Futures 3(3):035001
Overvelde JT, Weaver JC, Hoberman C, Bertoldi K (2017) Rational design of reconfigurable prismatic architected materials. Nature 541(7637):347–352
Hakimi O, Gelpi JL, Krallinger M, Curi F, Repchevsky D, Ginebra MP (2020) The devices, experimental scaffolds, and biomaterials ontology (DEB): a tool for mapping, annotation, and analysis of biomaterials’ data. Adv Func Mater 30(16):1909910
Kerner J, Dogan A, von Recum H (2021) Machine learning and big data provide crucial insight for future biomaterials discovery and research. Acta Biomater 130:54–65
Suwardi A, Wang F, Xue K, Han MY, Teo P, Wang P, Wang S, Liu Y, Ye E, Li Z, Loh XJ (2022) Machine learning-driven biomaterials evolution. Adv Mater 34(1):e2102703
Upadhya R, Kosuri S, Tamasi M, Meyer TA, Atta S, Webb MA, Gormley AJ (2021) Automation and data-driven design of polymer therapeutics. Adv Drug Deliv Rev 171:1–28
Vermeulen S, Honig F, Vasilevich A, Roumans N, Romero M, DedeEren A, Tuvshindorj U, Alexander M, Carlier A, Williams P, Uquillas J, de Boer J (2021) Expanding biomaterial surface topographical design space through natural surface reproduction. Adv Mater 33(31):e2102084
Cai J, Chu X, Xu K, Li H, Wei J (2020) Machine learning-driven new material discovery. Nanoscale Adv 2(8):3115–3130
Cencer MM, Moore JS, Assary RS (2022) Machine learning for polymeric materials: an introduction. Polym Int 71(5):537–542
Gao CC, Min X, Fang MH, Tao TY, Zheng XH, Liu YG, Wu XW, Huang ZH (2022) Innovative Materials science via machine learning. Adv Funct Mater. https://doi.org/10.1002/adfm.202108044
Juan YF, Dai YB, Yang Y, Zhang J (2021) Accelerating materials discovery using machine learning. J Mater Sci Technol 79:178–190
Morgan D, Jacobs R (2020) Opportunities and challenges for machine learning in materials science. In: Clarke DR (Ed.) Annu Rev Mater Res 50:71–103.
Saal JE, Oliynyk AO, Meredig B (2020) Machine learning in materials discovery: confirmed predictions and their underlying approaches. In: Clarke DR (Ed.) Annu Rev Mater Res 50: 49–69.
Sparks TD, Kauwe SK, Parry ME, Tehrani AM, Brgoch J (2020) Machine learning for structural materials. In: Clarke DR (Ed.) Annu Rev Mater Res 50: 27–48.
Geris L, Lambrechts T, Carlier A, Papantoniou I (2018) The future is digital: tissue engineering. Curr Opin Biomed Eng 6:92–98
Dobrzańska J, Dobrzański LB, Gołombek K, Dobrzański LA, Dobrzańska-Danikiewicz AD (2021) Virtual approach to the comparative analysis of biomaterials used in endodontic treatment. Processes 9(6):926
Echlin MP, Burnett TL, Polonsky AT, Pollock TM, Withers PJ (2020) Serial sectioning in the SEM for three dimensional materials science. Curr Opin Solid State Mater Sci 24(2):100817
YiWang W, Li J, Liu W, Liu Z-K (2019) Integrated computational materials engineering for advanced materials: a brief review. Comput Mater Sci 158:42–48
Basu B, Gowtham NH, Xiao Y, Kalidindi SR, Leong KW (2022) Biomaterialomics: data science-driven pathways to develop fourth-generation biomaterials. Acta Biomater 143:1–25
Kharmanda G (2023) Challenges and future perspectives for additively manufactured polylactic acid using fused filament fabrication in dentistry. J Funct Biomater 14(7):334
Sanz-Herrera JA, Reina-Romo E (2011) Cell-biomaterial mechanical interaction in the framework of tissue engineering: insights, computational modeling and perspectives. Int J Mol Sci 12(11):8217–8244
Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD (2007) Microfluidic scaffolds for tissue engineering. Nat Mater 6(11):908–915
Perier-Metz C, Duda GN, Checa S (2020) Mechano-biological computer model of scaffold-supported bone regeneration: effect of bone graft and scaffold structure on large bone defect tissue patterning. Front Bioeng Biotechnol 8:585799
Cai Y, Pan J, Li Z (2021) Mathematical modeling of intraplaque neovascularization and hemorrhage in a carotid atherosclerotic plaque. Research Square Platform LLC
Friedman JH (2002) Stochastic gradient boosting. Comput Stat Data Anal 38(4):367–378
Wu C, Fang JG, Zhang ZP, Entezari A, Sun GY, Swain MV, Li Q (2020) Fracture modeling of brittle biomaterials by the phase-field method. Eng Fract Mech 224:106752
Fang JG, Wu CQ, Rabczuk T, Wu C, Ma CG, Sun GY, Li Q (2019) Phase field fracture in elasto-plastic solids: abaqus implementation and case studies. Theoret Appl Fract Mech 103:102252
Fang JG, Wu CQ, Li J, Liu Q, Wu C, Sun GY, Li Q (2019) Phase field fracture in elasto-plastic solids: Variational formulation for multi-surface plasticity and effects of plastic yield surfaces and hardening. Int J Mech Sci 156:382–396
Cao Y, Karimi M, Kamrani E, Nourani P, Manesh AM, Momenieskandari H, Anqi AE (2021) Machine learning methods help accurate estimation of the hydrogen solubility in biomaterials. Int J Hydrogen Energy 47:3611–3624
Milad A, Hussein SH, Khekan AR, Rashid M, Al-Msari H, Tran TH (2022) Development of ensemble machine learning approaches for designing fiber-reinforced polymer composite strain prediction model. Eng Comput 38(4):3625–3637
Doan Tran H, Kim C, Chen L, Chandrasekaran A, Batra R, Venkatram S, Kamal D, Lightstone JP, Gurnani R, Shetty P, Ramprasad M, Laws J, Shelton M, Ramprasad R (2020) Machine-learning predictions of polymer properties with polymer genome. J Appl Phys. https://doi.org/10.1063/5.0023759
Li J, Gao H, Ye Z, Deng J, Ouyang D (2022) In silico formulation prediction of drug/cyclodextrin/polymer ternary complexes by machine learning and molecular modeling techniques. Carbohydr Polym 275:118712
Patel RA, Webb MA (2023) Data-driven design of polymer-based biomaterials: high-throughput simulation, experimentation, and machine learning. ACS Appl Bio Mater
Ueki Y, Seko N, Maekawa Y (2021) Machine learning approach for prediction of the grafting yield in radiation-induced graft polymerization. Appl Mater Today 25:101158
Choudhury A, Konnur T, Chattopadhyay PP, Pal S (2019) Structure prediction of multi-principal element alloys using ensemble learning. Eng Comput 37(3):1003–1022
Deng ZH, Yin HQ, Jiang X, Zhang C, Zhang GF, Xu B, Yang GQ, Zhang T, Wu M, Qu XH (2020) Machine-learning-assisted prediction of the mechanical properties of Cu-Al alloy. Int J Miner Metall Mater 27(3):362–373
Diao YP, Yan LC, Gao KW (2021) Improvement of the machine learning-based corrosion rate prediction model through the optimization of input features. Mater Design 198:109326
Goud VS, Rahul RM, Phanikumar G (2022) Prediction of growth velocity of undercooled multicomponent metallic alloys using a machine learning approach. Scripta Mater 207:114309
Nyshadham C, Rupp M, Bekker B, Shapeev AV, Mueller T, Rosenbrock CW, Csányi G, Wingate DW, Hart GLW (2019) Machine-learned multi-system surrogate models for materials prediction. NPJ Computat Mater. https://doi.org/10.1038/s41524-019-0189-9
Lupo Pasini M, Li YW, Yin J, Zhang J, Barros K, Eisenbach M (2021) Fast and stable deep-learning predictions of material properties for solid solution alloys. J Phys Condens Matter 33(8):084005
Qiao L, Lai ZH, Liu Y, Bao A, Zhu JC (2021) Modelling and prediction of hardness in multi-component alloys: a combined machine learning, first principles and experimental study. J Alloys Compd 853
Hou HB, Wang JF, Ye L, Zhu SJ, Wang LG, Guan SK (2023) Prediction of mechanical properties of biomedical magnesium alloys based on ensemble machine learning. Mater Lett 348:134605
Liao HC, Zhao BJ, Suo XJ, Wang QG (2019) Prediction models for macro shrinkage of aluminum alloys based on machine learning algorithms. Mater Today Commun 21
Huang XY, Wang H, Xue WH, Ullah A, Xiang S, Huang HL, Meng L, Ma G, Zhang GZ (2020) A combined machine learning model for the prediction of time-temperature-transformation diagrams of high-alloy steels. J Alloys Compd 823
Klimenko D, Stepanov N, Li J, Fang Q, Zherebtsov S (2021) Machine learning-based strength prediction for refractory high-entropy alloys of the Al-Cr-Nb-Ti-V-Zr System. Materials 14(23):7213
Machaka R, Motsi GT, Raganya LM, Radingoana PM, Chikosha S (2021) Machine learning-based prediction of phases in high-entropy alloys: a data article. Data Brief 38:107346
Liu XD, Li X, He QF, Liang DD, Zhou ZQ, Ma J, Yang Y, Shen J (2020) Machine learning-based glass formation prediction in multicomponent alloys. Acta Mater 201:182–190
Ward L, O’Keeffe SC, Stevick J, Jelbert GR, Aykol M, Wolverton C (2018) A machine learning approach for engineering bulk metallic glass alloys. Acta Mater 159:102–111
Sadeghian Dehkord E, Kerckhofs G, Compere P, Lambert F, Geris L (2023) An empirical model linking physico-chemical biomaterial characteristics to intra-oral bone formation. J Funct Biomater 14(7):388
Wang Z, Dabaja R, Chen L, Banu M (2023) Machine learning unifies flexibility and efficiency of spinodal structure generation for stochastic biomaterial design. Sci Rep 13(1):5414
Yuan RH, Tian Y, Xue DZ, Xue DQ, Zhou YM, Ding XD, Sun J, Lookman T (2019) Accelerated search for BaTiO-based ceramics with large energy storage at low fields using machine learning and experimental design. Adv Sci 6(21):1901395
Yu JW, Wang Y, Dai ZQ, Yang FM, Fallahpour A, Nasiri-Tabrizi B (2021) Structural features modeling of substituted hydroxyapatite nanopowders as bone fillers via machine learning. Ceram Int 47(7):9034–9047
Verma D, Dong Y, Sharma M, Chaudhary AK (2022) Advanced processing of 3D printed biocomposite materials using artificial intelligence. Mater Manuf Processes 37(5):518–538
MasoodChaudry U, Hamad K, Abuhmed T (2021) Machine learning-aided design of aluminum alloys with high performance. Mater Today Commun 26:101897
Liang TT, Wang JS, Xue CP, Zhang C, Zhang MS (2022) Design of high strength and electrically conductive aluminium alloys by machine learning. Mater Sci Technol 38(2):116–129
Dong RZ, Dan YB, Li X, Hu JJ (2021) Inverse design of composite metal oxide optical materials based on deep transfer learning and global optimization. Comput Mater Sci 188:110166
Fatehi E, Sarvestani HY, Ashrafi B, Akbarzadeh AH (2021) Accelerated design of architectured ceramics with tunable thermal resistance via a hybrid machine learning and finite element approach. Mater Design 210:110056
Furtado C, Pereira LF, Tavares RP, Salgado M, Otero F, Catalanotti G, Arteiro A, Bessa MA, Camanho PP (2021) A methodology to generate design allowables of composite laminates using machine learning. Int J Solids Struct 233:111095
Gao H, Zhong S, Zhang W, Igou T, Berger E, Reid E, Zhao Y, Lambeth D, Gan L, Afolabi MA, Tong Z, Lan G, Chen Y (2022) Revolutionizing membrane design using machine learning-bayesian optimization. Environ Sci Technol 56(4):2572–2581
Huang XY, Jin C, Zhang C, Zhang H, Fu HW (2021) Machine learning assisted modelling and design of solid solution hardened high entropy alloys. Mater Design 211:110177
Jeon J, Seo N, Kim HJ, Lee MH, Lim HK, Son SB, Lee SJ (2021) Inverse design of Fe-based bulk metallic glasses using machine learning. Metals 11(5):729
Kim C, Batra R, Chen LH, Tran H, Ramprasad R (2021) Polymer design using genetic algorithm and machine learning. Comput Mater Sci 186:110067
Lai F, Sun Z, Saji SE, He Y, Yu X, Zhao H, Guo H, Yin Z (2021) Machine learning-aided crystal facet rational design with ionic liquid controllable synthesis. Small 17(12):e2100024
Liu Y, Zhang D, Tang Y, Zhang Y, Chang Y, Zheng J (2021) Machine learning-enabled design and prediction of protein resistance on self-assembled monolayers and beyond. ACS Appl Mater Interfaces 13(9):11306–11319
Pei Z, Rozman KA, Dogan ON, Wen Y, Gao N, Holm EA, Hawk JA, Alman DE, Gao MC (2021) Machine-learning microstructure for inverse material design. Adv Sci 8(23):e2101207
Tu D, Yan J, Xie Y, Li J, Feng S, Xia M, Li J, Leung AP (2022) Accelerated design for magnetocaloric performance in Mn-Fe-P-Si compounds using machine learning. J Mater Sci Technol 96:241–247
Wang C, Wei X, Ren D, Wang X, Xu W (2022) High-throughput map design of creep life in low-alloy steels by integrating machine learning with a genetic algorithm. Mater Design 213:110326
Tao QL, Lu T, Sheng Y, Li L, Lu WC, Li MJ (2021) Machine learning aided design of perovskite oxide materials for photocatalytic water splitting. J Energy Chem 60:351–359
Zhang HT, Fu HD, Zhu SC, Yong W, Xie JX (2021) Machine learning assisted composition effective design for precipitation strengthened copper alloys. Acta Mater 215:117118
Zhang JY, Li YW, Zhao TY, Zhang Q, Zuo L, Zhang K (2021) Machine-learning based design of digital materials for elastic wave control. Extreme Mech Lett 48:101372
Gandhi A, Hasan MMF (2022) Machine learning for the design and discovery of zeolites and porous crystalline materials. Curr Opin Chem Eng 35:100739
Khalvandi A, Tayebi L, Kamarian S, Saber-Samandari S, Song JI (2023) Data-driven supervised machine learning to predict the compressive response of porous PVA/Gelatin hydrogels and in-vitro assessments: employing design of experiments. Int J Biol Macromol 253(Pt 3):126906
Fang Z, Zhang M, Wang H, Chen J, Yuan H, Wang M, Ye S, Jia YG, Sheong FK, Wang Y, Wang L (2023) Marriage of high-throughput gradient surface generation with statistical learning for the rational design of functionalized biomaterials. Adv Mater 35(49):e2303253
Alber M, Tepole AB, Cannon WR, De S, Dura-Bernal S, Garikipati K, Karniadakis G, Lytton WW, Perdikaris P, Petzold L, Kuhl E (2019) Integrating machine learning and multiscale modeling-perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. NPJ Digit Med 2(1):115
Zhao YC, Zhang Y, Jiang F, Wu C, Wan B, Syeda R, Li Q, Shen B, Ju LA (2023) A novel computational biomechanics framework to model vascular mechanopropagation in deep bone marrow. Adv Healthc Mater 12(8):e2201830
Halilaj E, Rajagopal A, Fiterau M, Hicks JL, Hastie TJ, Delp SL (2018) Machine learning in human movement biomechanics: best practices, common pitfalls, and new opportunities. J Biomech 81:1–11
Bes A, Rao S, Pandya HJ (2019) Engineering approaches for characterizing soft tissue mechanical properties: a review. Clin Biomech 69:127–140
Galbusera F, Casaroli G, Bassani T (2019) Artificial intelligence and machine learning in spine research. JOR Spine 2(1):e1044
Phellan R, Hachem B, Clin J, Mac-Thiong JM, Duong L (2021) Real-time biomechanics using the finite element method and machine learning: review and perspective. Med Phys 48(1):7–18
Mouloodi S, Rahmanpanah H, Gohari S, Burvill C, Tse KM, Davies HMS (2021) What can artificial intelligence and machine learning tell us? A review of applications to equine biomechanical research. J Mech Behav Biomed Mater 123:104728
Matijevich ES, Scott LR, Volgyesi P, Derry KH, Zelik KE (2020) Combining wearable sensor signals, machine learning and biomechanics to estimate tibial bone force and damage during running. Hum Mov Sci 74:102690
Derie R, Robberechts P, Van den Berghe P, Gerlo J, De Clercq D, Segers V, Davis J (2020) Tibial acceleration-based prediction of maximal vertical loading rate during overground running: a machine learning approach. Front Bioeng Biotechnol 8:33
Robberechts P, Derie R, Van den Berghe P, Gerlo J, De Clercq D, Segers V, Davis J (2021) Predicting gait events from tibial acceleration in rearfoot running: a structured machine learning approach. Gait Posture 84:87–92
Dorschky E, Nitschke M, Martindale CF, van den Bogert AJ, Koelewijn AD, Eskofier BM (2020) CNN-based estimation of sagittal plane walking and running biomechanics from measured and simulated inertial sensor data. Front Bioeng Biotechnol 8:604
Stetter BJ, Ringhof S, Krafft FC, Sell S, Stein T (2019) Estimation of knee joint forces in sport movements using wearable sensors and machine learning. Sensors 19(17):3690
Ardestani MM, Chen ZX, Wang L, Lian Q, Liu YX, He JK, Li DC, Jin ZM (2014) Feed forward artificial neural network to predict contact force at medial knee joint: application to gait modification. Neurocomputing 139:114–129
Nicholson KF, Richardson RT, van Roden EAR, Quinton RG, Anzilotti KF, Richards JG (2019) Machine learning algorithms for predicting scapular kinematics. Med Eng Phys 65:39–45
Bouteraa Y, Abdallah IB, Boukthir K (2023) A new wrist-forearm rehabilitation protocol integrating human biomechanics and SVM-based machine learning for muscle fatigue estimation. Bioengineering 10(2):219
Mundt M, Born Z, Goldacre M, Alderson J (2022) Estimating ground reaction forces from two-dimensional pose data: a biomechanics-based comparison of alphapose, blazepose, and openpose. Sensors 23(1):78
McCabe MV, Van Citters DW, Chapman RM (2023) Hip joint angles and moments during stair ascent using neural networks and wearable sensors. Bioengineering 10(7):784
Babu A, Ranpariya S, Sinha DK, Mandal D (2023) Deep learning enabled perceptive wearable sensor: an interactive gadget for tracking movement disorder. Adv Mater Technol 8(14):2300046
Qiu JG, Li Y, Liu HQ, Lin S, Pang L, Sun G, Song YZ (2023) Research on motion recognition based on multi-dimensional sensing data and deep learning algorithms. Math Biosci Eng 20(8):14578–14595
Luo Y, Li J, He K, Cheuk W (2022) A hierarchical attention-based method for sleep staging using movement and cardiopulmonary signals. IEEE J Biomed Health Inform 27(3):1354–1363
Mani N, Haridoss P, George B (2023) Smart suspenders with sensors and machine learning for human activity monitoring. IEEE Sens J 23(9):10159–10167
Zong KQ, Wang Y, Zhao YP, Zhang LX (2023) Background separation of sports athletes and motion image analysis based on skeleton segmentation algorithm. Soft Comput. https://doi.org/10.1007/s00500-023-08830-5
Yoo KS (2023) Motion prediction using brain waves based on artificial intelligence deep learning recurrent neural network. J Exerc Rehabil 19(4):219–227
M.A.A. Faisal, S. Mahmud, M.E.H. Chowdhury, A. Khandakar, M.U. Ahmed, A. Alqahtani, M. Alhatou, Robust and novel attention guided MultiResUnet model for 3D ground reaction force and moment prediction from foot kinematics, Neural Computing & Applications (2023) 1–17.
Tang C, Chen X, Gong J, Occhipinti LG, Gao S (2022) WMNN: wearables-based multi-column neural network for human activity recognition. IEEE J Biomed Health Inform 27(1):339–350
Jannat MKA, Islam MS, Yang SH, Liu H (2023) Efficient Wi-Fi-based human activity recognition using adaptive antenna elimination. IEEE Access 11:105440–105454
Zhang ZJ, Kan EC (2023) Novel muscle sensing by radiomyography (RMG) and its application to hand gesture recognition. IEEE Sens J 23(17):20116–20128
Zhang J, Zhao Y, Shone F, Li Z, Frangi AF, Xie SQ, Zhang Z-Q (2022) Physics-informed deep learning for musculoskeletal modeling: predicting muscle forces and joint kinematics from surface EMG. IEEE Trans Neural Syst Rehabil Eng 31:484–493
Mokhtari-Jafari F, Amoabediny G, Dehghan MM (2020) Role of biomechanics in vascularization of tissue-engineered bones. J Biomech 110:109920
Carniel EL, Toniolo I, Fontanella CG (2020) Computational biomechanics: in silico tools for the investigation of surgical procedures and devices. Bioengineering 7(2):48
Santolini E, West R, Giannoudis PV (2015) Risk factors for long bone fracture non-union: a stratification approach based on the level of the existing scientific evidence. Injury 46(Suppl 8):S8–S19
Langerhuizen DW, Janssen SJ, Mallee WH, Van Den Bekerom MP, Ring D, Kerkhoffs GM, Jaarsma RL, Doornberg JN (2019) What are the applications and limitations of artificial intelligence for fracture detection and classification in orthopaedic trauma imaging? A systematic review. Clin Orthop Relat Res 477(11):2482
Bromiley PA, Clark EM, Poole KE (2020) Computer-aided diagnostic systems for osteoporotic vertebral fracture detection: opportunities and challenges. J Bone Miner Res 35(12):2305–2306
Shen SC, Pena Fernandez M, Tozzi G, Buehler MJ (2021) Deep learning approach to assess damage mechanics of bone tissue. J Mech Behav Biomed Mater 123:104761
Pranata YD, Wang KC, Wang JC, Idram I, Lai JY, Liu JW, Hsieh IH (2019) Deep learning and SURF for automated classification and detection of calcaneus fractures in CT images. Comput Methods Programs Biomed 171:27–37
Tanzi L, Vezzetti E, Moreno R, Aprato A, Audisio A, Masse A (2020) Hierarchical fracture classification of proximal femur X-ray images using a multistage deep learning approach. Eur J Radiol 133:109373
Erne F, Dehncke D, Herath SC, Springer F, Pfeifer N, Eggeling R, Kuper MA (2023) Deep learning in the detection of rare fractures—development of a “deep learning convolutional network” model for detecting acetabular fractures. Z Orthop Unfall 161(1):42–50
Whittier DE, Samelson EJ, Hannan MT, Burt LA, Hanley DA, Biver E, Szulc P, Sornay-Rendu E, Merle B, Chapurlat R, Lespessailles E, Wong AKO, Goltzman D, Khosla S, Ferrari S, Bouxsein ML, Kiel DP, Boyd SK (2022) Bone microarchitecture phenotypes identified in older adults are associated with different levels of osteoporotic fracture risk. J Bone Miner Res 37(3):428–439
Reid S, Schousboe JT, Kimelman D, Monchka BA, Jafari Jozani M, Leslie WD (2021) Machine learning for automated abdominal aortic calcification scoring of DXA vertebral fracture assessment images: a pilot study. Bone 148:115943
Niculescu B, Faur CI, Tataru T, Diaconu BM, Cruceru M (2020) Investigation of biomechanical characteristics of orthopedic implants for tibial plateau fractures by means of deep learning and support vector machine classification. Appl Sci 10(14):4697
Ergün GB, Güney S (2021) Classification of canine maturity and bone fracture time based on X-ray images of long bones. IEEE Access 9:109004–109011
Bagaria R, Wadhwani S, Wadhwani AK (2021) Bone fractures detection using support vector machine and error backpropagation neural network. Optik 247:168021
Villamor E, Monserrat C, Del Rio L, Romero-Martin JA, Ruperez MJ (2020) Prediction of osteoporotic hip fracture in postmenopausal women through patient-specific FE analyses and machine learning. Comput Methods Programs Biomed 193:105484
Lucchinetti E, Stussi E (2004) Prediction of elasticity constants in small biomaterial samples such as bone: a comparison between classical optimization techniques and identification with artificial neural networks. Proc Inst Mech Eng H 218(6):389–405
Nazemi SM, Amini M, Kontulainen SA, Milner JS, Holdsworth DW, Masri BA, Wilson DR, Johnston JD (2017) Optimizing finite element predictions of local subchondral bone structural stiffness using neural network-derived density-modulus relationships for proximal tibial subchondral cortical and trabecular bone. Clin Biomech 41:1–8
Vukicevic AM, Jovicic GR, Jovicic MN, Milicevic VL, Filipovic ND (2018) Assessment of cortical bone fracture resistance curves by fusing artificial neural networks and linear regression. Comput Methods Biomech Biomed Engin 21(2):169–176
Rahmanpanah H, Mouloodi S, Burvill C, Gohari S, Davies HMS (2020) Prediction of load-displacement curve in a complex structure using artificial neural networks: a study on a long bone. Int J Eng Sci 154:103319
Mouloodi S, Rahmanpanah H, Burvill C, Gohari S, Davies HMS (2021) Experimental, regression learner, numerical, and artificial neural network analyses on a complex composite structure subjected to compression loading. Mech Adv Mater Struct 29(17):2437–2453
Christopher JJ, Ramakrishnan S (2008) Assessment and classification of mechanical strength components of human femur trabecular bone using texture analysis and neural network. J Med Syst 32(2):117–122
Deng B, Tan KBC, Lu Y, Zaw K, Zhang J, Liu GR, Geng JP (2009) Inverse identification of elastic modulus of dental implant-bone interfacial tissue using neural network and FEA model. Inverse Prob Sci Eng 17(8):1073–1083
Baseri H, Rabiee SM, Moztarzadeh F, Solati-Hashjin M (2010) Mechanical strength and setting times estimation of hydroxyapatite cement by using neural network. Mater Des 31(5):2585–2591
Hambli R (2011) Apparent damage accumulation in cancellous bone using neural networks. J Mech Behav Biomed Mater 4(6):868–878
Campoli G, Weinans H, Zadpoor AA (2012) Computational load estimation of the femur. J Mech Behav Biomed Mater 10:108–119
Zadpoor AA, Campoli G, Weinans H (2013) Neural network prediction of load from the morphology of trabecular bone. Appl Math Model 37(7):5260–5276
Barkaoui A, Chamekh A, Merzouki T, Hambli R, Mkaddem A (2014) Multiscale approach including microfibril scale to assess elastic constants of cortical bone based on neural network computation and homogenization method. Int J Numer Method Biomed Eng 30(3):318–338
Garijo N, Martínez J, García-Aznar JM, Pérez MA (2014) Computational evaluation of different numerical tools for the prediction of proximal femur loads from bone morphology. Comput Methods Appl Mech Eng 268:437–450
Garijo N, Verdonschot N, Engelborghs K, Garcia-Aznar JM, Perez MA (2017) Subject-specific musculoskeletal loading of the tibia: computational load estimation. J Mech Behav Biomed Mater 65:334–343
Khaterchi H, Chamekh A, BelHadjSalah H (2015) Artificial neural network analysis for modeling fibril structure in bone. Int J Precis Eng Manuf 16(3):581–587
Khovanova NA, Shaikhina T, Mallick KK (2015) Neural networks for analysis of trabecular bone in osteoarthritis. Bioinspired Biomimetic Nanobiomater 4(1):90–100
Barkaoui A, Tlili B, Vercher-Martinez A, Hambli R (2016) A multiscale modelling of bone ultrastructure elastic proprieties using finite elements simulation and neural network method. Comput Methods Programs Biomed 134:69–78
Taylor M, Perilli E, Martelli S (2017) Development of a surrogate model based on patient weight, bone mass and geometry to predict femoral neck strains and fracture loads. J Biomech 55:121–127
Mouloodi S, Rahmanpanah H, Burvill C, Davies HMS (2020) Prediction of load in a long bone using an artificial neural network prediction algorithm. J Mech Behav Biomed Mater 102:103527
Mouloodi S, Rahmanpanah H, Burvill C, Davies HMS (2020) Prediction of displacement in the equine third metacarpal bone using a neural network prediction algorithm. Biocybern Biomed Eng 40(2):849–863
Mouss ME, Zellagui S, Nasraoui M, Hambli R (2020) Parametric investigation of the effects of load level on fatigue crack growth in trabecular bone based on artificial neural network computation. Proc Inst Mech Eng H 234(8):784–793
Zhong J, Shibata Y, Wu C, Watanabe C, Chen J, Zheng K, Hu J, Swain MV, Li Q (2023) Functional non-uniformity of periodontal ligaments tunes mechanobiological stimuli across soft- and hard-tissue interfaces. Acta Biomater 170:240–249
Berni M, Veronesi F, Fini M, Giavaresi G, Marchiori G, Oliviero F (2023) Relations between structure/composition and mechanics in osteoarthritic regenerated articular tissue: a machine learning approach. Int J Mol Sci 24(17):13374
Disney CM, Lee PD, Hoyland JA, Sherratt MJ, Bay BK (2018) A review of techniques for visualising soft tissue microstructure deformation and quantifying strain Ex Vivo. J Microsc 272(3):165–179
Xi W, Saw TB, Delacour D, Lim CT, Ladoux B (2018) Material approaches to active tissue mechanics. Nat Rev Mater 4(1):23–44
Liang L, Liu M, Martin C, Elefteriades JA, Sun W (2017) A machine learning approach to investigate the relationship between shape features and numerically predicted risk of ascending aortic aneurysm. Biomech Model Mechanobiol 16(5):1519–1533
Madani A, Bakhaty A, Kim J, Mubarak Y, Mofrad MRK (2019) Bridging finite element and machine learning modeling: stress prediction of arterial walls in atherosclerosis. J Biomech Eng. https://doi.org/10.1115/1.4043290
Liang L, Liu M, Martin C, Sun W (2018) A machine learning approach as a surrogate of finite element analysis-based inverse method to estimate the zero-pressure geometry of human thoracic aorta. Int J Numer Method Biomed Eng 34(8):e3103
Tonutti M, Gras G, Yang GZ (2017) A machine learning approach for real-time modelling of tissue deformation in image-guided neurosurgery. Artif Intell Med 80:39–47
Liang L, Liu M, Sun W (2017) A deep learning approach to estimate chemically-treated collagenous tissue nonlinear anisotropic stress-strain responses from microscopy images. Acta Biomater 63:227–235
Nguyen-Le DH, Ballit A, Dao TT (2023) A novel deep learning-driven approach for predicting the pelvis soft-tissue deformations toward a real-time interactive childbirth simulation. Eng Appl Artif Intell 126:107150
Dalton D, Husmeier D, Gao H (2023) Physics-informed graph neural network emulation of soft-tissue mechanics. Comput Methods Appl Mech Eng 417:116351
Cilla M, Perez-Rey I, Martinez MA, Pena E, Martinez J (2018) On the use of machine learning techniques for the mechanical characterization of soft biological tissues. Int J Numer Method Biomed Eng 34(10):e3121
Liu M, Liang L, Sun W (2019) Estimation of in vivo constitutive parameters of the aortic wall using a machine learning approach. Comput Methods Appl Mech Eng 347:201–217
Gasser TC, Ogden RW, Holzapfel GA (2006) Hyperelastic modelling of arterial layers with distributed collagen fibre orientations. J R Soc Interface 3(6):15–35
Wittek A, Derwich W, Karatolios K, Fritzen CP, Vogt S, Schmitz-Rixen T, Blase C (2016) A finite element updating approach for identification of the anisotropic hyperelastic properties of normal and diseased aortic walls from 4D ultrasound strain imaging. J Mech Behav Biomed Mater 58:122–138
Wittek A, Karatolios K, Bihari P, Schmitz-Rixen T, Moosdorf R, Vogt S, Blase C (2013) In vivo determination of elastic properties of the human aorta based on 4D ultrasound data. J Mech Behav Biomed Mater 27:167–183
Gonzalez-Saiz E, Garcia-Gonzalez D (2023) Model-driven identification framework for optimal constitutive modeling from kinematics and rheological arrangement. Comput Methods Appl Mech Eng 415:116211
Meyer GV (1867) Die architektur der spongiosa, Archisfur Anatomie, Physiologie und wissenschaftliche Medizin. Reichert und DuBois-Reymonds Archiv 34:615–628
Mehlenbacher RD, Kolbl R, Lay A, Dionne JA (2017) Nanomaterials for in vivo imaging of mechanical forces and electrical fields. Nat Rev Mater 3(2):1–17
Hadjidakis DJ, Androulakis II (2006) Bone remodeling. Ann NY Acad Sci 1092(1):385–396
Fazzalari NL (2011) Bone fracture and bone fracture repair. Osteoporos Int 22(6):2003–2006
Li Z, Muller R, Ruffoni D (2018) Bone remodeling and mechanobiology around implants: Insights from small animal imaging. J Orthop Res 36(2):584–593
Della Corte A, Giorgio I, Scerrato D (2020) A review of recent developments in mathematical modeling of bone remodeling. Proc Inst Mech Eng H 234(3):273–281
Cohen DO, Aboutaleb SMG, Johnson AW, Norato JA (2021) Bone adaptation-driven design of periodic scaffolds. J Mech Design. https://doi.org/10.1115/1.4050928
Perier-Metz C, Duda GN, Checa S (2021) Initial mechanical conditions within an optimized bone scaffold do not ensure bone regeneration—an in silico analysis. Biomech Model Mechanobiol 20(5):1723–1731
Metz C, Duda GN, Checa S (2020) Towards multi-dynamic mechano-biological optimization of 3D-printed scaffolds to foster bone regeneration. Acta Biomater 101:117–127
Sturm S, Zhou S, Mai YW, Li Q (2010) On stiffness of scaffolds for bone tissue engineering-a numerical study. J Biomech 43(9):1738–1744
Schouman T, Schmitt M, Adam C, Dubois G, Rouch P (2016) Influence of the overall stiffness of a load-bearing porous titanium implant on bone ingrowth in critical-size mandibular bone defects in sheep. J Mech Behav Biomed Mater 59:484–496
Sanz-Herrera JA, Garía-Aznar JM, Doblaré M (2008) Micro-macro numerical modelling of bone regeneration in tissue engineering. Comput Methods Appl Mech Eng 197(33–40):3092–3107
Sanz-Herrera JA, Garcia-Aznar JM, Doblare M (2009) On scaffold designing for bone regeneration: a computational multiscale approach. Acta Biomater 5(1):219–229
Chen Y, Zhou S, Li Q (2011) Microstructure design of biodegradable scaffold and its effect on tissue regeneration. Biomaterials 32(22):5003–5014
Chang C-C, Chen Y, Zhou S, Mai Y-W, Li Q (2016) Computational design for scaffold tissue engineering, springer series in biomaterials science and engineering. Springer, Berlin Heidelberg, pp 349–369
Bashkuev M, Checa S, Postigo S, Duda G, Schmidt H (2015) Computational analyses of different intervertebral cages for lumbar spinal fusion. J Biomech 48(12):3274–3282
Checa S, Prendergast PJ (2009) A mechanobiological model for tissue differentiation that includes angiogenesis: a lattice-based modeling approach. Ann Biomed Eng 37(1):129–145
Checa S, Prendergast PJ (2010) Effect of cell seeding and mechanical loading on vascularization and tissue formation inside a scaffold: a mechano-biological model using a lattice approach to simulate cell activity. J Biomech 43(5):961–968
Sandino C, Checa S, Prendergast PJ, Lacroix D (2010) Simulation of angiogenesis and cell differentiation in a CaP scaffold subjected to compressive strains using a lattice modeling approach. Biomaterials 31(8):2446–2452
Kelly DJ, Prendergast PJ (2005) Mechano-regulation of stem cell differentiation and tissue regeneration in osteochondral defects. J Biomech 38(7):1413–1422
Cheong VS, Blunn GW, Coathup MJ, Fromme P (2018) A novel adaptive algorithm for 3D finite element analysis to model extracortical bone growth. Comput Methods Biomech Biomed Engin 21(2):129–138
Huiskes R, Van Driel WD, Prendergast PJ, Soballe K (1997) A biomechanical regulatory model for periprosthetic fibrous-tissue differentiation. J Mater Sci Mater Med 8(12):785–788
Prendergast PJ, Huiskes R, Soballe K (1997) ESB Research Award 1996: biophysical stimuli on cells during tissue differentiation at implant interfaces. J Biomech 30(6):539–548
Sanz-Herrera JA, Garcia-Aznar JM, Doblare M (2008) A mathematical model for bone tissue regeneration inside a specific type of scaffold. Biomech Model Mechanobiol 7(5):355–366
Ghosh R, Chanda S, Chakraborty D (2021) Qualitative predictions of bone growth over optimally designed macro-textured implant surfaces obtained using NN-GA based machine learning framework. Med Eng Phys 95:64–75
Kilarski WW, Samolov B, Petersson L, Kvanta A, Gerwins P (2009) Biomechanical regulation of blood vessel growth during tissue vascularization. Nat Med 15(6):657–664
Han Y, Huang K, Yao QP, Jiang ZL (2018) Mechanobiology in vascular remodeling. Natl Sci Rev 5(6):933–946
Adamo L, Naveiras O, Wenzel PL, McKinney-Freeman S, Mack PJ, Gracia-Sancho J, Suchy-Dicey A, Yoshimoto M, Lensch MW, Yoder MC, Garcia-Cardena G, Daley GQ (2009) Biomechanical forces promote embryonic haematopoiesis. Nature 459(7250):1131–1135
Thondapu V, Bourantas CV, Foin N, Jang IK, Serruys PW, Barlis P (2017) Biomechanical stress in coronary atherosclerosis: emerging insights from computational modelling. Eur Heart J 38(2):81–92
Pratt SJP, Lee RM, Martin SS (2020) The mechanical microenvironment in breast cancer. Cancers 12(6):1452
Petho Z, Najder K, Bulk E, Schwab A (2019) Mechanosensitive ion channels push cancer progression. Cell Calcium 80:79–90
Lee G, Han SB, Lee JH, Kim HW, Kim DH (2019) Cancer mechanobiology: microenvironmental sensing and metastasis. ACS Biomater Sci Eng 5(8):3735–3752
Petridou NI, Spiro Z, Heisenberg CP (2017) Multiscale force sensing in development. Nat Cell Biol 19(6):581–588
Gillespie PG, Walker RG (2001) Molecular basis of mechanosensory transduction. Nature 413(6852):194–202
Nemec S, Lam J, Zhong J, Heu C, Timpson P, Li Q, Youkhana J, Sharbeen G, Phillips PA, Kilian KA (2021) Interfacial curvature in confined coculture directs stromal cell activity with spatial corralling of pancreatic cancer cells. Adv Biol 5(6):e2000525
Dattatrey M, Tiwari AK, Ghoshal B, Singh J (2019) Predicting bone modeling parameters in response to mechanical loading. IEEE Access 7:122561–122572
Tiwari AK, Kumar N (2018) Establishing the relationship between loading parameters and bone adaptation. Med Eng Phys 56:16–26
Bonnevie ED, Ashinsky BG, Dekky B, Volk SW, Smith HE, Mauck RL (2021) Cell morphology and mechanosensing can be decoupled in fibrous microenvironments and identified using artificial neural networks. Sci Rep 11(1):5950
Guo ZY, Peng XQ, Moran B (2006) A composites-based hyperelastic constitutive model for soft tissue with application to the human annulus fibrosus. J Mech Phys Solids 54(9):1952–1971
Wang X, Schoen JA, Rentschler ME (2013) A quantitative comparison of soft tissue compressive viscoelastic model accuracy. J Mech Behav Biomed Mater 20:126–136
Simon BR (1992) Multiphase poroelastic finite element models for soft tissue structures. Appl Mech Rev 45(6):191–218
Gordon S, Pluddemann A (2017) Tissue macrophages: heterogeneity and functions. BMC Biol 15(1):53
Zhong J, Pierantoni M, Weinkamer R, Brumfeld V, Zheng K, Chen J, Swain MV, Weiner S, Li Q (2021) Microstructural heterogeneity of the collagenous network in the loaded and unloaded periodontal ligament and its biomechanical implications. J Struct Biol 213(3):107772
Bourgat JF (1979) Numerical experiments of the homogenization method. Lecture notes in mathematics. Springer, Berlin Heidelberg, pp 330–356
Li Z (2021) Predicting bone regeneration from machine learning. Nat Comput Sci 1(8):509–510
Liu LZ, Shi Q, Chen Q, Li ZY (2019) Mathematical modeling of bone in-growth into undegradable porous periodic scaffolds under mechanical stimulus. J Tissue Eng 10:2041731419827167
Shi Q, Shui HT, Chen Q, Li ZY (2020) How does mechanical stimulus affect the coupling process of the scaffold degradation and bone formation: an approach. Comput Biol Med 117:103588
Zahedmanesh H, Lally C (2012) A multiscale mechanobiological modelling framework using agent-based models and finite element analysis: application to vascular tissue engineering. Biomech Model Mechanobiol 11(3–4):363–377
Kelkel J, Surulescu C (2012) A multiscale approach to cell migration in tissue networks. Math Models Methods Appl Sci. https://doi.org/10.1142/S0218202511500175
Holden EC, Collis J, Brook BS, O’Dea RD (2018) A multiphase multiscale model for nutrient limited tissue growth. Anziam Journal 59(4):499–532
Blaszczyk M, Hackl K (2022) Multiscale modeling of cancellous bone considering full coupling of mechanical, electric and magnetic effects. Biomech Model Mechanobiol 21(1):163–187
Zhou M, Bezci SE, O’Connell GD (2020) Multiscale composite model of fiber-reinforced tissues with direct representation of sub-tissue properties. Biomech Model Mechanobiol 19(2):745–759
Marino M, Vairo G (2014) Stress and strain localization in stretched collagenous tissues via a multiscale modelling approach. Comput Methods Biomech Biomed Engin 17(1):11–30
Nikpasand M, Mahutga RR, Bersie-Larson LM, Gacek E, Barocas VH (2021) A hybrid microstructural-continuum multiscale approach for modeling hyperelastic fibrous soft tissue. J Elast 145(1–2):295–319
Hadi MF, Sander EA, Barocas VH (2012) Multiscale model predicts tissue-level failure from collagen fiber-level damage. J Biomech Eng 134(9):091005
Heltai L, Caiazzo A, Muller LO (2021) Multiscale coupling of one-dimensional vascular models and elastic tissues. Ann Biomed Eng 49(12):3243–3254
Heltai L, Caiazzo A (2019) Multiscale modeling of vascularized tissues via nonmatching immersed methods. Int J Numer Method Biomed Eng 35(12):e3264
Dalbosco M, Carniel TA, Fancello EA, Holzapfel GA (2021) Multiscale numerical analyses of arterial tissue with embedded elements in the finite strain regime. Comput Methods Appl Mech Eng 381:113844
Peng L, Trucu D, Lin P, Thompson A, Chaplain MA (2017) A multiscale mathematical model of tumour invasive growth. Bull Math Biol 79(3):389–429
Chakrabarti A, Verbridge S, Stroock AD, Fischbach C, Varner JD (2012) Multiscale models of breast cancer progression. Ann Biomed Eng 40(11):2488–2500
Sansalone V, Lemaire T, Naili S (2007) Modélisation multi-échelle des propriétés mécaniques de l’os: étude à l’échelle de la fibrille. Comptes Rendus Mécanique 335(8):436–442
Ghanbari J, Naghdabadi R (2009) Nonlinear hierarchical multiscale modeling of cortical bone considering its nanoscale microstructure. J Biomech 42(10):1560–1565
Li JF, Li SG (2016) Multiscale models of compact bone. Int J Biomath 9(3)
Kwon YW, Clumpner BR (2018) Multiscale modeling of human bone. Multiscale Multidisc Model Exp Design 1(2):133–143
Sanz-Herrera JA, Mora-Macías J, Reina-Romo E, Domínguez J, Doblaré M (2019) Multiscale characterisation of cortical bone tissue. Appl Sci 9(23):5228
Kaiser J, Lemaire T, Naili S, Sansalone V (2009) Modèle multi-échelle du transport de fluide dans un milieu poreux chargé avec échanges cationiques: application aux tissus osseux. Comptes Rendus Mécanique 337(11–12):768–775
Perrin E, Bou-Said B, Massi F (2019) Numerical modeling of bone as a multiscale poroelastic material by the homogenization technique. J Mech Behav Biomed Mater 91:373–382
Ilic S, Hackl K, Gilbert R (2010) Application of the multiscale FEM to the modeling of cancellous bone. Biomech Model Mechanobiol 9(1):87–102
Huang ZQ, Nie YF, Li YQ (2020) Microstructural modeling and multiscale mechanical properties analysis of cancellous bone. Comput Mater Continua 62(1):245–265
Goncalves Coelho P, Rui Fernandes P, Carrico Rodrigues H (2011) Multiscale modeling of bone tissue with surface and permeability control. J Biomech 44(2):321–329
Colloca M, Blanchard R, Hellmich C, Ito K, van Rietbergen B (2014) A multiscale analytical approach for bone remodeling simulations: linking scales from collagen to trabeculae. Bone 64:303–313
Zhao F, Vaughan TJ, McNamara LM (2015) Multiscale fluid-structure interaction modelling to determine the mechanical stimulation of bone cells in a tissue engineered scaffold. Biomech Model Mechanobiol 14(2):231–243
Pastrama MI, Scheiner S, Pivonka P, Hellmich C (2018) A mathematical multiscale model of bone remodeling, accounting for pore space-specific mechanosensation. Bone 107:208–221
Causin P, Sacco R, Verri M (2013) A multiscale approach in the computational modeling of the biophysical environment in artificial cartilage tissue regeneration. Biomech Model Mechanobiol 12(4):763–780
Sharma A, Molla S, Katti KS, Katti DR (2017) Multiscale models of degradation and healing of bone tissue engineering nanocomposite scaffolds. J Nanomech Micromech. https://doi.org/10.1061/(ASCE)NM.2153-5477.0000133
Adouni M, Mbarki R, Al-Khatib F, Eilaghi A (2021) Multiscale modeling of knee ligament biomechanics. Int J Numer Method Biomed Eng 37(1):e3413
Hambli R (2011) Numerical procedure for multiscale bone adaptation prediction based on neural networks and finite element simulation. Finite Elem Anal Des 47(7):835–842
Hambli R, Katerchi H, Benhamou CL (2011) Multiscale methodology for bone remodelling simulation using coupled finite element and neural network computation. Biomech Model Mechanobiol 10(1):133–145
Pled F, Desceliers C, Zhang TY (2021) A robust solution of a statistical inverse problem in multiscale computational mechanics using an artificial neural network. Comput Methods Appl Mech Eng 373:113540
Hashemi MS, Baniassadi M, Baghani M, George D, Remond Y, Sheidaei A (2020) A novel machine learning based computational framework for homogenization of heterogeneous soft materials: application to liver tissue. Biomech Model Mechanobiol 19(3):1131–1142
Mora-Macías J, Ayensa-Jiménez J, Reina-Romo E, Doweidar MH, Domínguez J, Doblaré M, Sanz-Herrera JA (2020) A multiscale data-driven approach for bone tissue biomechanics. Comput Methods Appl Mech Eng 368:113136
Kirchdoerfer T, Ortiz M (2016) Data-driven computational mechanics. Comput Methods Appl Mech Eng 304:81–101
Kirchdoerfer T, Ortiz M (2018) Data-driven computing in dynamics. Int J Numer Meth Eng 113(11):1697–1710
Arjoca S, Robu A, Neagu M, Neagu A (2023) Mathematical and computational models in spheroid-based biofabrication. Acta Biomater 165:125–139
Deckers T, Hall GN, Papantoniou I, Aerts JM, Bloemen V (2022) A platform for automated and label-free monitoring of morphological features and kinetics of spheroid fusion. Front Bioeng Biotechnol 10:946992
Park SY, Kim SJ, Park CH, Kim J, Lee DY (2023) Data-driven prediction models for forecasting multistep ahead profiles of mammalian cell culture toward bioprocess digital twins. Biotechnol Bioeng 120(9):2494–2508
Hasnain A, Balakrishnan S, Joshy DM, Smith J, Haase SB, Yeung E (2023) Learning perturbation-inducible cell states from observability analysis of transcriptome dynamics. Nat Commun 14(1):3148
Wu C, Wan B, Entezari A, Fang J, Xu Y, Li Q (2024) Machine learning-based design for additive manufacturing in biomedical engineering. Int J Mech Sci 266:108828
Goh GD, Sing SL, Yeong WY (2021) A review on machine learning in 3D printing: applications, potential, and challenges. Artif Intell Rev 54(1):63–94
Meng LB, McWilliams B, Jarosinski W, Park HY, Jung YG, Lee J, Zhang J (2020) Machine learning in additive manufacturing: a review. Jom 72(6):2363–2377
Qi XB, Chen GF, Li Y, Cheng X, Li CP (2019) Applying neural-network-based machine learning to additive manufacturing: current applications, challenges and future perspectives. Engineering 5(4):721–729
Aghajani S, Wu C, Li Q, Fang J (2023) Additively manufactured composite lattices: a state-of-the-art review on fabrications, architectures, constituent materials, mechanical properties, and future directions. Thin-Walled Struct 197:111539
Groll J, Boland T, Blunk T, Burdick JA, Cho DW, Dalton PD, Derby B, Forgacs G, Li Q, Mironov VA, Moroni L, Nakamura M, Shu W, Takeuchi S, Vozzi G, Woodfield TB, Xu T, Yoo JJ, Malda J (2016) Biofabrication: reappraising the definition of an evolving field. Biofabrication 8(1):013001
Wang Y, Cui H, Esworthy T, Mei D, Wang Y, Zhang LG (2022) Emerging 4D printing strategies for next-generation tissue regeneration and medical devices. Adv Mater 34(20):e2109198
Zhou M, Hou J, Zhang G, Luo C, Zeng Y, Mou S, Xiao P, Zhong A, Yuan Q, Yang J, Wang Z, Sun J (2019) Tuning the mechanics of 3D-printed scaffolds by crystal lattice-like structural design for breast tissue engineering. Biofabrication 12(1):015023
Li JH, Wu CT, Chu PK, Gelinsky M (2020) 3D printing of hydrogels: Rational design strategies and emerging biomedical applications. Mater Sci Eng R-Rep 140:100543
Walker BW, Lara RP, Mogadam E, Yu CH, Kimball W, Annabi N (2019) Rational design of microfabricated electroconductive hydrogels for biomedical applications. Prog Polym Sci 92:135–157
Wu C, Luo JJ, Zhong JX, Xu YA, Wan BY, Huang WW, Fang JG, Steven GP, Sun GY, Li Q (2023) Topology optimisation for design and additive manufacturing of functionally graded lattice structures using derivative-aware machine learning algorithms. Addit Manuf 78:103833
Tian CX, Li TJ, Bustillos J, Bhattacharya S, Turnham T, Yeo JJ, Moridi A (2021) Data-driven approaches toward smarter additive manufacturing. Adv Intell Syst 3(12):2100014
Almesmari A, Alagha AN, Naji MM, Sheikh-Ahmad J, Jarrar F (2023) Recent advancements in design optimization of lattice-structured materials. Adv Eng Mater 25(17):2201780
Wu YZ, Fang JG, Wu C, Li CY, Sun GY, Li Q (2023) Additively manufactured materials and structures: a state-of-the-art review on their mechanical characteristics and energy absorption. Int J Mech Sci 246:108102
E. Wang, R. Yao, Q. Li, X. Hu, G. Sun, Lightweight metallic cellular materials: a systematic review on mechanical characteristics and engineering applications, International Journal of Mechanical Sciences (2023) 108795.
Baykasoğlu A, Baykasoğlu C, Cetin E (2020) Multi-objective crashworthiness optimization of lattice structure filled thin-walled tubes. Thin-Walled Struct 149:106630
Song J, Zhou WZ, Wang YJ, Fan R, Wang YC, Chen JY, Lu Y, Li LX (2019) Octet-truss cellular materials for improved mechanical properties and specific energy absorption. Mater Des 173:107773
Tafazoli M, Nouri MD (2022) Experimental and numerical study and multi-objective optimisation of quasi-static compressive test on three-dimensional printed lattice-core sandwich structures. Int J Crashworthiness 27(1):117–127
Vijayavenkataraman S, Zhang L, Zhang S, HsiFuh JY, Lu WF (2018) Triply periodic minimal surfaces sheet scaffolds for tissue engineering applications: an optimization approach toward biomimetic scaffold design. ACS Appl Bio Mater 1(2):259–269
Beyer C, Figueroa D (2016) Design and analysis of lattice structures for additive manufacturing. J Manuf Sci Eng Trans Asme. https://doi.org/10.1115/1.4033957
Fernandes MC, Aizenberg J, Weaver JC, Bertoldi K (2021) Mechanically robust lattices inspired by deep-sea glass sponges. Nat Mater 20(2):237–241
Nian Y, Wan S, Li X, Su Q, Li M (2019) How does bio-inspired graded honeycomb filler affect energy absorption characteristics? Thin-Walled Struct 144:106269
Xie Y, Bai HL, Liu ZH, Chen NN (2020) A novel bionic structure inspired by luffa sponge and its cushion properties. Appl Sci 10(7):2584
Meena K, Singamneni S (2019) A new auxetic structure with significantly reduced stress concentration effects. Mater Des 173:107779
Wang CC, Yang KC, Lin KH, Wu CC, Liu YL, Lin FH, Chen IH (2014) A biomimetic honeycomb-like scaffold prepared by flow-focusing technology for cartilage regeneration. Biotechnol Bioeng 111(11):2338–2348
Yang K, Xu SQ, Shen JH, Zhou SW, Xie YM (2016) Energy absorption of thin-walled tubes with pre-folded origami patterns: numerical simulation and experimental verification. Thin-Walled Struct 103:33–44
Fang J, Sun G, Qiu N, Kim NH, Li Q (2016) On design optimization for structural crashworthiness and its state of the art. Struct Multidiscip Optim 55(3):1091–1119
Biswas JK, Dey S, Karmakar SK, Roychowdhury A, Datta S (2020) Design of patient specific spinal implant (pedicle screw fixation) using fe analysis and soft computing techniques. Curr Med Imaging 16(4):371–382
Chatterjee S, Dey S, Majumder S, RoyChowdhury A, Datta S (2019) Computational intelligence based design of implant for varying bone conditions. Int J Numer Method Biomed Eng 35(6):e3191
Roy S, Dey S, Khutia N, RoyChowdhury A, Datta S (2018) Design of patient specific dental implant using FE analysis and computational intelligence techniques. Appl Soft Comput 65:272–279
Roy S, Dey S, Khutia N, Chowdhury AR, Datta S (2018) Design of patient specific dental implant using FE analysis and computational intelligence techniques. Appl Soft Comput 65:272–279
Wu C, Fang JG, Zhou SW, Zhang ZP, Sun GY, Steven GP, Li Q (2021) A path-dependent level set topology optimization with fracture criterion. Comput Struct 249:106515
Cucinotta F, Guglielmino E, Longo G, Risitano G, Santonocito D, Sfravara F (2019) Topology optimization additive manufacturing-oriented for a biomedical application. Advances on mechanics, design engineering and manufacturing II. Springer, pp 184–193
Xiao DM, Yang YQ, Su XB, Wang D, Luo ZY (2012) Topology optimization of microstructure and selective laser melting fabrication for metallic biomaterial scaffolds. Trans Nonferrous Metals Soc China 22(10):2554–2561
Poh PSP, Valainis D, Bhattacharya K, van Griensven M, Dondl P (2019) Optimization of bone scaffold porosity distributions. Sci Rep 9(1):9170
Lin CY, Kikuchi N, Hollister SJ (2004) A novel method for biomaterial scaffold internal architecture design to match bone elastic properties with desired porosity. J Biomech 37(5):623–636
Hollister SJ, Lin CY (2007) Computational design of tissue engineering scaffolds. Comput Methods Appl Mech Eng 196(31–32):2991–2998
Chen Y, Schellekens M, Zhou S, Cadman J, Li W, Appleyard R, Li Q (2011) Design optimization of scaffold microstructures using wall shear stress criterion towards regulated flow-induced erosion. J Biomech Eng 133(8):081008
Zhang Z, Chen J, Li E, Li W, Swain M, Li Q (2016) Topological design of all-ceramic dental bridges for enhancing fracture resistance. Int J Numer Method Biomed Eng 32(6):e02749
Wu C, Fang JG, Zhou SW, Zhang ZP, Sun GY, Steven GP, Li Q (2020) Level-set topology optimization for maximizing fracture resistance of brittle materials using phase-field fracture model. Int J Numer Meth Eng 121(13):2929–2945
Park J, Lee D, Sutradhar A (2019) Topology optimization of fixed complete denture framework. Int J Numer Method Biomed Eng 35(6):e3193
Gupta Y, Iyer R, Dommeti VK, Nutu E, Rana M, Merdji A, Biswas JK, Roy S (2021) Design of dental implant using design of experiment and topology optimization: a finite element analysis study. Proc Inst Mech Eng H 235(2):157–166
Chang CL, Chen CS, Huang CH, Hsu ML (2012) Finite element analysis of the dental implant using a topology optimization method. Med Eng Phys 34(7):999–1008
Xue H, Saha SC, Beier S, Jepson N, Luo Z (2021) Topological optimization of auxetic coronary stents considering hemodynamics. Front Bioeng Biotechnol 9:728914
Xue H, Luo Z, Brown T, Beier S (2020) Design of self-expanding auxetic stents using topology optimization. Front Bioeng Biotechnol 8:736
James KA, Waisman H (2016) Layout design of a bi-stable cardiovascular stent using topology optimization. Comput Methods Appl Mech Eng 305:869–890
Farhan Khan M, Brackett D, Ashcroft I, Tuck C, Wildman R (2017) A novel approach to design lesion-specific stents for minimum recoil. J Med Devices. https://doi.org/10.1115/1.4034880
Al-Tamimi AA, Huang BY, Vyas C, Hernandez M, Peach C, Bartolo P (2019) Topology optimised metallic bone plates produced by electron beam melting: a mechanical and biological study. Int J Adv Manuf Technol 104(1–4):195–210
Al-Tamimi AA (2021) 3D topology optimization and mesh dependency for redesigning locking compression plates aiming to reduce stress shielding. Int J Bioprint 7(3):339
Kumar KES, Rakshit S (2020) Topology optimization of the hip bone for gait cycle. Struct Multidiscip Optim 62(4):2035–2049
Deng XW, Wang YJ, Yan JH, Liu T, Wang ST (2016) Topology optimization of total femur structure: application of parameterized level set method under geometric constraints. J Mech Design. https://doi.org/10.1115/1.4031803
Zhong ZC, Wei SH, Wang JP, Feng CK, Chen CS, Yu CH (2006) Finite element analysis of the lumbar spine with a new cage using a topology optimization method. Med Eng Phys 28(1):90–98
Chuah HG, Abd Rahim I, Yusof MI (2010) Topology optimisation of spinal interbody cage for reducing stress shielding effect. Comput Methods Biomech Biomed Engin 13(3):319–326
Xu YN, Gao YK, Wu C, Fang JG, Sun GY, Steven GP, Li Q (2021) On design of carbon fiber reinforced plastic (CFRP) laminated structure with different failure criteria. Int J Mech Sci 196:106251
Xu YA, Gao YK, Wu C, Fang JG, Li Q (2019) Robust topology optimization for multiple fiber-reinforced plastic (FRP) composites under loading uncertainties. Struct Multidiscip Optim 59(3):695–711
Wu C, Gao YK, Fang JG, Lund E, Li Q (2019) Simultaneous discrete topology optimization of ply orientation and thickness for carbon fiber reinforced plastic-laminated structures. J Mech Design. https://doi.org/10.1115/1.4042222
Wu C, Gao YK, Fang JG, Lund E, Li Q (2017) Discrete topology optimization of ply orientation for a carbon fiber reinforced plastic (CFRP) laminate vehicle door. Mater Des 128:9–19
Wu C, Fang JG, Li Q (2019) Multi-material topology optimization for thermal buckling criteria. Comput Methods Appl Mech Eng 346:1136–1155
Hu C, Fang XQ, Huang WH (2008) Multiple scattering of shear waves and dynamic stress from a circular cavity buried in a semi-infinite slab of functionally graded materials. Eng Fract Mech 75(5):1171–1183
Bendsøe MP, Sigmund O (1999) Material interpolation schemes in topology optimization. Arch Appl Mech (Ingenieur Archiv) 69(9–10):635–654
Wang MY, Wang XM, Guo DM (2003) A level set method for structural topology optimization. Comput Methods Appl Mech Eng 192(1–2):227–246
Allaire G, Jouve F, Toader AM (2004) Structural optimization using sensitivity analysis and a level-set method. J Comput Phys 194(1):363–393
Liu C, Zhu YC, Sun Z, Li DD, Du ZL, Zhang WS, Guo X (2018) An efficient moving morphable component (MMC)-based approach for multi-resolution topology optimization. Struct Multidiscip Optim 58(6):2455–2479
Yu Y, Hur T, Jung J, Jang IG (2018) Deep learning for determining a near-optimal topological design without any iteration. Struct Multidiscip Optim 59(3):787–799
Nie ZG, Lin T, Jiang HL, Kara LB (2021) TopologyGAN: topology optimization using generative adversarial networks based on physical fields over the initial domain. J Mech Design. https://doi.org/10.1115/1.4049533
Chandrasekhar A, Suresh K (2021) TOuNN: topology optimization using neural networks. Struct Multidiscip Optim 63(3):1135–1149
White DA, Arrighi WJ, Kudo J, Watts SE (2019) Multiscale topology optimization using neural network surrogate models. Comput Methods Appl Mech Eng 346:1118–1135
Cang RJ, Yao H, Ren Y (2019) One-shot generation of near-optimal topology through theory-driven machine learning. Comput Aided Des 109:12–21
Lynch ME, Sarkar S, Maute K (2019) Machine learning to aid tuning of numerical parameters in topology optimization. J Mech Design. https://doi.org/10.1115/1.4044228
Behzadi MM, Ilies HT (2021) GANTL: towards practical and real-time topology optimization with conditional GANs and transfer learning. J Mech Des 144(2):1–32
Kallioras NA, Lagaros ND (2021) MLGen: generative design framework based on machine learning and topology optimization. Appl Sci 11(24):12044
Yamasaki S, Yaji K, Fujita K (2021) Data-driven topology design using a deep generative model. Struct Multidiscip Optim 64(3):1401–1420
Wang DL, Xiang C, Pan Y, Chen AR, Zhou XY, Zhang YQ (2022) A deep convolutional neural network for topology optimization with perceptible generalization ability. Eng Optim 54(6):973–988
Zheng S, He ZZ, Liu HL (2021) Generating three-dimensional structural topologies via a U-Net convolutional neural network. Thin-Walled Struct 159:107263
Seo J, Kapania RK (2023) Topology optimization with advanced CNN using mapped physics-based data. Struct Multidiscip Optim 66(1):21
Abueidda DW, Koric S, Sobh NA (2020) Topology optimization of 2D structures with nonlinearities using deep learning. Comput Struct 237:106283
Chandrasekhar A, Suresh K (2021) Multi-material topology optimization using neural networks. Comput Aid Design 136:103017
Bielecki D, Patel D, Rai R, Dargush GF (2021) Multi-stage deep neural network accelerated topology optimization. Struct Multidiscip Optim 64(6):3473–3487
Vulimiri PS, Deng H, Dugast F, Zhang X, To AC (2021) Integrating geometric data into topology optimization via neural style transfer. Materials 14(16)
Kallioras NA, Kazakis G, Lagaros ND (2020) Accelerated topology optimization by means of deep learning. Struct Multidiscip Optim 62(3):1185–1212
Liu Z, Xia L, Xia Q, Shi TL (2020) Data-driven design approach to hierarchical hybrid structures with multiple lattice configurations. Struct Multidiscip Optim 61(6):2227–2235
Chi H, YY Zhang, Tang TLE, Mirabella L, Dalloro L, Song, L, Paulino GH (2021) Universal machine learning for topology optimization. Comput Methods Appl Mech Eng 375
Guo YL, Hsi JFY, Feng LW (2021) Multiscale topology optimisation with nonparametric microstructures using three-dimensional convolutional neural network (3D-CNN) models. Virtual Phys Prototyp 16(3):306–317
Kim C, Lee J, Yoo J (2021) Machine learning-combined topology optimization for functionary graded composite structure design. Comput Methods Appl Mech Eng 387
Qian C, Ye WJ (2021) Accelerating gradient-based topology optimization design with dual-model artificial neural networks. Struct Multidiscip Optim 63(4):1687–1707
Xia ZH, Zhang HB, Zhuang Z, Yu C, Yu JG, Gao L (2023) A machine-learning framework for isogeometric topology optimization. Struct Multidiscip Optim 66(4):83
Wang CP, Yao S, Wang ZJ, Hu J (2021) Deep super-resolution neural network for structural topology optimization. Eng Optim 53(12):2108–2121
Rade J, Balu A, Herron E, Pathak J, Ranade R, Sarkar S, Krishnamurthy A (2021) Algorithmically-consistent deep learning frameworks for structural topology optimization. Eng App Artif Intell 106:104483
Xue L, Liu J, Wen GL, Wang HX (2021) Efficient, high-resolution topology optimization method based on convolutional neural networks. Front Mech Eng 16(1):80–96
Ye HL, Li JC, Yuan BS, Wei N, Sui YK (2021) Acceleration design for continuum topology optimization by using Pix2pix neural network. Int J Appl Mech 13(4):2150042
Zhang ZY, Li Y, Zhou WE, Chen XQ, Yao W, Zhao Y (2021) TONR: An exploration for a novel way combining neural network with topology optimization. Comput Methods Appl Mech Eng 386:114083
Deng C, Wang Y, Qin C, Fu Y, Lu W (2022) Self-directed online machine learning for topology optimization. Nat Commun 13(1):388
Huo SL, Du BX, Zhao Y, Liu H, Shi YA, Chen XQ (2023) Thermal design of functionally graded cellular structures with multiple microstructure configurations through topology optimization. Compos Struct 313:116922
Zhang WS, Wang Y, Du ZL, Liu C, Youn SK, Guo X (2023) Machine-learning assisted topology optimization for architectural design with artistic flavor. Comput Methods Appl Mech Eng 413:116041
Jiang XC, Wang H, Li Y, Mo KJ (2020) Machine Learning based parameter tuning strategy for MMC based topology optimization. Adv Eng Softw 149:102841
Lei X, Liu C, Du ZL, Zhang WS, Guo X (2019) Machine learning-driven real-time topology optimization under moving morphable component-based framework. J Appl Mech Trans Asme. https://doi.org/10.1115/1.4041319
Zheng S, Fan HJ, Zhang ZY, Tian ZQ, Jia K (2021) Accurate and real-time structural topology prediction driven by deep learning under moving morphable component-based framework. Appl Math Model 97:522–535
Hoang VN, Nguyen NL, Tran DQ, Vu QV, Nguyen-Xuan H (2022) Data-driven geometry-based topology optimization. Struct Multidiscip Optim. https://doi.org/10.1007/s00158-022-03170-8
Kim KH, Yoon GH (2022) Acoustic topology optimization using moving morphable components in neural network-based design. Struct Multidiscip Optim. https://doi.org/10.1007/s00158-021-03137-1
Guo YH, Du ZL, Wang LB, Meng W, Zhang T, Su RY, Yang DS, Tang S, Guo X (2023) Data-driven topology optimization (DDTO) for three-dimensional continuum structures. Struct Multidiscip Optim 66(5):104
Du ZL, Luo JC, Xu Z, Jiang ZH, Ding XG, Cui TC, Guo X (2023) Higher-order topological insulators by ML-enhanced topology optimization. Int J Mech Sci 255:108441
Tucci M, Barmada S, Sani L, Thomopulos D, Fontana N (2019) IEEE, deep neural networks based surrogate model for topology optimization of electromagnetic devices. In: 2019 International applied computational electromagnetics society symposium (ACES)
Qiu C, Du SY, Yang JL (2021) A deep learning approach for efficient topology optimization based on the element removal strategy. Mater Design 212:110179
Deng H, To AC (2021) A parametric level set method for topology optimization based on deep neural network. J Mech Design. https://doi.org/10.1115/1.4050105
Liu H, Qi YT, Chen LX, Li YW, Xiao WL (2024) An efficient data-driven optimization framework for designing graded cellular structures. Appl Math Model 125:574–598
Xu YA, Gao YK, Wu C, Fang JG, Sun GY, Steven GP, Li Q (2021) Machine learning based topology optimization of fiber orientation for variable stiffness composite structures. Int J Numer Meth Eng 122(22):6736–6755
Hamdia KM, Ghasemi H, Bazi Y, AlHichri H, Alajlan N, Rabczuk T (2019) A novel deep learning based method for the computational material design of flexoelectric nanostructures with topology optimization. Finite Elem Anal Des 165:21–30
Huang MC, Cui TC, Liu C, Du ZL, Zhang JM, He CH, Guo X (2023) A Problem-Independent Machine Learning (PIML) enhanced substructure-based approach for large-scale structural analysis and topology optimization of linear elastic structures. Extreme Mech Lett 63:102041
Li YZ, Ge WJ, Liu B, Wang Z, Wang CM, Zhang YH (2023) Collaborative optimization of composite structure and fiber orientation through real-time correction of deep neural network (DNN) models with elite samples. Mech Adv Mater Struct. https://doi.org/10.1080/15376494.2023.2253018
Li M, Jia GF, Cheng ZB, Shi ZF (2021) Generative adversarial network guided topology optimization of periodic structures via subset simulation. Compos Struct 260:113254
Wang L, van Beek A, Da D, Chan Y-C, Zhu P, Chen W (2022) Data-driven multiscale design of cellular composites with multiclass microstructures for natural frequency maximization. Compos Struct 280:114949
Yaji K, Yamasaki S, Fujita K (2022) Data-driven multifidelity topology design using a deep generative model: application to forced convection heat transfer problems. Comput Methods Appl Mech Eng 388:114284
Zhou Y, Zhan HF, Zhang WH, Zhu JH, Bai JS, Wang QX, Gu YT (2020) A new data-driven topology optimization framework for structural optimization. Comput Struct 239:106310
Han SH, Han Q, Jiang TJ, Li CL (2023) Inverse design of phononic crystals for anticipated wave propagation by integrating deep learning and semi-analytical approach. Acta Mech 234(10):4879–4897
Hammond J, Pietropaoli M, Montomoli F (2022) Topology optimisation of turbulent flow using data-driven modelling. Struct Multidiscip Optim. https://doi.org/10.1007/s00158-021-03150-4
Jeong H, Batuwatta-Gamage C, Bai JS, Xie YM, Rathnayaka C, Zhou Y, Gu YT (2023) A complete physics-informed neural network-based framework for structural topology optimization. Comput Methods Appl Mech Eng 417:116401
Oh MK, Yoo J (2023) Functionally graded structure design for magnetic field applications. Comput Methods Appl Mech Eng 411:116057
Liu JK, Gaynor AT, Chen SK, Kang Z, Suresh K, Takezawa A, Li L, Kato J, Tang JY, Wang CCL, Cheng L, Liang X, To AC (2018) Current and future trends in topology optimization for additive manufacturing. Struct Multidiscip Optim 57(6):2457–2483
Xiong YL, Yao S, Zhao ZL, Xie YM (2020) A new approach to eliminating enclosed voids in topology optimization for additive manufacturing. Addit Manuf 32:101006
Azam FI, Rani AMA, Razak MAHA, Ali S, Aliyu AaA (2021) Additive manufacturing processes, challenges and applications: a review. Progress Eng Technol 3:93–111
Entezari A, Liu NC, Roohani I, Zhang Z, Chen J, Sarrafpour B, Zoellner H, Behi M, Zreiqat H, Li Q (2020) On design for additive manufacturing (DAM) parameter and its effects on biomechanical properties of 3D printed ceramic scaffolds. Mater Today Commun 23:101065
Zou J, Zhang YC, Feng ZY (2021) Topology optimization for additive manufacturing with self-supporting constraint. Struct Multidiscip Optim 63(5):2341–2353
Zhou L, Zhang WH (2019) Topology optimization method with elimination of enclosed voids. Struct Multidiscip Optim 60(1):117–136
Wang C, Xu B, Meng QX, Rong JH, Zhao YH (2020) Numerical performance of Poisson method for restricting enclosed voids in topology optimization. Comput Struct 239:106337
Li QH, Chen WJ, Liu ST, Tong LY (2016) Structural topology optimization considering connectivity constraint. Struct Multidiscip Optim 54(4):971–984
Gaynor AT, Guest JK (2016) Topology optimization considering overhang constraints: eliminating sacrificial support material in additive manufacturing through design. Struct Multidiscip Optim 54(5):1157–1172
Wang YG, Gao JC, Kang Z (2018) Level set-based topology optimization with overhang constraint: towards support-free additive manufacturing. Comput Methods Appl Mech Eng 339:591–614
Garaigordobil A, Ansola R, Santamaria J, de Bustos IF (2018) A new overhang constraint for topology optimization of self-supporting structures in additive manufacturing. Struct Multidiscip Optim 58(5):2003–2017
Allaire G, Jakabcin L (2018) Taking into account thermal residual stresses in topology optimization of structures built by additive manufacturing. Math Models Methods Appl Sci 28(12):2313–2366
Zhou MD, Liu YC, Lin ZQ (2019) Topology optimization of thermal conductive support structures for laser additive manufacturing. Comput Methods Appl Mech Eng 353:24–43
Misiun G, van de Ven E, Langelaar M, Geijselaers H, van Keulen F, van den Boogaard T, Ayas C (2021) Topology optimization for additive manufacturing with distortion constraints. Comput Methods Appl Mech Eng 386:114095
Cheng L, Liang X, Bai JX, Chen Q, Lemon J, To A (2019) On utilizing topology optimization to design support structure to prevent residual stress induced build failure in laser powder bed metal additive manufacturing. Addit Manuf 27:290–304
Xu S, Liu J, Ma Y (2022) Residual stress constrained self-support topology optimization for metal additive manufacturing. Comput Methods Appl Mech Eng 389:114380
Pellens J, Lombaert G, Michiels M, Craeghs T, Schevenels M (2020) Topology optimization of support structure layout in metal-based additive manufacturing accounting for thermal deformations. Struct Multidiscip Optim 61(6):2291–2303
Miki T, Yamada T (2021) Topology optimization considering the distortion in additive manufacturing. Finite Elements Analy Design 193:103558
Tan RK, Zhang NL, Ye W (2019) A deep learning–based method for the design of microstructural materials. Struct Multidiscip Optim 61(4):1417–1438
Weiss BM, Hamel JM, Ganter MA, Storti DW (2021) Data-driven additive manufacturing constraints for topology optimization. J Manuf Sci Eng Trans ASME. https://doi.org/10.1115/1.4048264
Li C, Liu ZY, Fang XY, Guo YB (2018) Residual stress in metal additive manufacturing. 4th Cirp Conf Surf Integrity 71:348–353
Paul R, Anand S, Gerner F (2014) Effect of thermal deformation on part errors in metal powder based additive manufacturing processes. J Manuf Sci Eng Trans ASME. https://doi.org/10.1115/1.4026524
Choi Y-H, Kim C-M, Jeong H-S, Youn J-H (2016) Influence of bed temperature on heat shrinkage shape error in fdm additive manufacturing of the abs-engineering plastic. World J Eng Technol 04(03):186–192
Bresson Y, Tongne A, Selva P, Arnaud L (2022) Numerical modelling of parts distortion and beam supports breakage during selective laser melting (SLM) additive manufacturing. Int J Adv Manuf Technol 119(9–10):5727–5742
Safronov VA, Khmyrov RS, Kotoban DV, Gusarov AV (2017) Distortions and residual stresses at layer-by-layer additive manufacturing by fusion. J Manuf Sci Eng—Trans ASME 139(3):031017
Cao XF, Jiang YB, Zhao T, Wang PD, Wang YZ, Chen ZH, Li Y, Xiao DB, Fang DN (2020) Compression experiment and numerical evaluation on mechanical responses of the lattice structures with stochastic geometric defects originated from additive-manufacturing. Compos Part B—Eng 194:108030
Liu BS, Jiang C, Li GY, Huang XD (2020) Topology optimization of structures considering local material uncertainties in additive manufacturing. Comput Methods Appl Mech Eng 360:112786
Davoudinejad A, Diaz-Perez LC, Quagliotti D, Pedersen DB, Albajez-García JA, Yagüe-Fabra JA, Tosello G (2018) Additive manufacturing with vat polymerization method for precision polymer micro components production. 15th Cirp Conf Comput Aid Tolerancing, Cirp Cat 2018 75:98–102
Nath P, Olson JD, Mahadevan S, Lee YT (2020) Optimization of fused filament fabrication process parameters under uncertainty to maximize part geometry accuracy. Addit Manuf. https://doi.org/10.1016/j.addma.2020.101331
Li ZS, Wang L, Lv TQ (2023) Additive manufacturing-oriented concurrent robust topology optimization considering size control. Int J Mech Sci 250:108269
Thillaithevan D, Bruce P, Santer M (2022) Robust multiscale optimization accounting for spatially-varying material uncertainties. Struct Multidiscip Optim 65(2):40
Entezari A, Liu N-C, Zhang Z, Fang J, Wu C, Wan B, Swain M, Li Q (2023) Nondeterministic multiobjective optimization of 3D printed ceramic tissue scaffolds. J Mech Behav Biomed Mater 138:105580
Zhang YM, Karnati S, Nag S, Johnson N, Khan G, Ribic B (2022) Accelerating additive design with probabilistic machine learning. ASCE-ASME J Risk Uncertainty Eng Syst Part B. https://doi.org/10.1115/1.4051699
Wang Z, Liu PW, Xiao YH, Cui XY, Hui Z, Chen L (2019) A data-driven approach for process optimization of metallic additive manufacturing under uncertainty. J Manuf Sci Eng Trans ASME. https://doi.org/10.1115/1.4043798
Guo SH, Agarwal M, Cooper C, Tian Q, Gao RBX, Grace WG, Guo Y (2022) Machine learning for metal additive manufacturing: towards a physics-informed data-driven paradigm. J Manuf Syst 62:145–163
Kumar S, Gopi T, Harikeerthana N, Gupta MK, Gaur V, Krolczyk GM, Wu C (2023) Machine learning techniques in additive manufacturing: a state of the art review on design, processes and production control. J Intell Manuf 34(1):21–55
de Souza Borges Ferreira R, Sabbaghi A, Huang Q (2020) Automated geometric shape deviation modeling for additive manufacturing systems via bayesian neural networks. IEEE Trans Autom Sci Eng 17(2):584–598
Huang Q, Wang YX, Lyu M, Lin WZ (2020) Shape deviation generator—a convolution framework for learning and predicting 3-D printing shape accuracy. IEEE Trans Autom Sci Eng 17(3):1486–1500
Alacoque L, Watkins RT, Tamijani AY (2021) Stress-based and robust topology optimization for thermoelastic multi-material periodic microstructures. Comput Methods Appl Mech Eng 379:113749
Gao Q, Liao WH (2021) Energy absorption of thin walled tube filled with gradient auxetic structures-theory and simulation. Int J Mech Sci 201:106475
Hong H, Hu M, Dai L (2020) dynamic mechanical behavior of hierarchical resin honeycomb by 3D printing. Polymers 13(1):19
Liang YJ, Zhou W, Liu YJ, Li ZS, Yang Y, Xi HF, Wu ZG (2021) Energy absorption and deformation behavior of 3D printed triply periodic minimal surface stainless steel cellular structures under compression. Steel Res Int. https://doi.org/10.1002/srin.202000411
Rahman H, Yarali E, Zolfagharian A, Serjouei A, Bodaghi M (2021) Energy absorption and mechanical performance of functionally graded soft-hard lattice structures. Materials 14(6):1366
Bujny M, Olhofer M, Aulig N, Duddeck F (2021) Topology optimization of 3D-printed joints under crash loads using evolutionary algorithms. Struct Multidiscip Optim 64(6):4181–4206
Andrew JJ, Schneider J, Ubaid J, Velmurugan R, Gupta NK, Kumar S (2021) Energy absorption characteristics of additively manufactured plate-lattices under low- velocity impact loading. Int J Impact Eng 149:103768
Wang L, Xia HJ, Yang YW, Cai YR, Qiu ZP (2019) A novel approach of reliability-based topology optimization for continuum structures under interval uncertainties. Rapid Prototyp J 25(9):1455–1474
Schaedler TA, Ro CJ, Sorensen AE, Eckel Z, Yang SS, Carter WB, Jacobsen AJ (2014) Designing metallic microlattices for energy absorber applications. Adv Eng Mater 16(3):276–283
Chatterjee T, Chakraborty S, Goswami S, Adhikari S, Friswell MI (2021) Robust topological designs for extreme metamaterial micro-structures. Sci Rep 11(1):15221
Keshavarzzadeh V, James KA (2019) Robust multiphase topology optimization accounting for manufacturing uncertainty via stochastic collocation. Struct Multidiscip Optim 60(6):2461–2476
Zhang HR, Zhou H, Zhou ZX, Zeng HZ, Zhang XY, Yang JZ, Lei HS, Han FS (2021) Energy absorption diagram characteristic of metallic self-supporting 3D lattices fabricated by additive manufacturing and design method of energy absorption structure. Int J Solids Struct 226:111082
Jin N, Yan ZY, Wang YW, Cheng HW, Zhang HM (2021) Effects of heat treatment on microstructure and mechanical properties of selective laser melted Ti-6Al-4V lattice materials. Int J Mech Sci 190:106042
Wu Y, Yang L (2021) Elastic and failure characteristics of additive manufactured thin wall lattice structures with defects. Thin-Walled Struct 161:107493
Yang J (2017) Design and engineering of metamaterials and metasurfaces: from fundamentals to applications. In: Proceedings of the 3rd world congress on mechanical, chemical, and material engineering. Avestia Publishing
Wang CY, Vangelatos Z, Grigoropoulos CP, Ma Z (2022) Micro-engineered architected metamaterials for cell and tissue engineering. Mater Today Adv 13:100206
Martinez-Marquez D, Delmar Y, Sun S, Stewart RA (2020) Exploring macroporosity of additively manufactured titanium metamaterials for bone regeneration with quality by design: a systematic literature review. Materials 13(21):4794
Dogan E, Bhusal A, Cecen B, Miri AK (2020) 3D printing metamaterials towards tissue engineering. Appl Mater Today 20:100752
Zheng L, Kumar S, Kochmann DM (2021) Data-driven topology optimization of spinodoid metamaterials with seamlessly tunable anisotropy. Comput Methods Appl Mech Eng 383:113894
Wang LW, Tao SY, Zhu P, Chen W (2021) Data-driven topology optimization with multiclass microstructures using latent variable Gaussian process. J Mech Design. https://doi.org/10.1115/1.4048628
Wang LW, Chan YC, Liu Z, Zhu P, Chen W (2020) Data-driven metamaterial design with Laplace-Beltrami spectrum as “shape-DNA.” Struct Multidiscip Optim 61(6):2613–2628
Wang LW, Chan YC, Ahmed F, Liu Z, Zhu P, Chen W (2020) Deep generative modeling for mechanistic-based learning and design of metamaterial systems. Comput Methods Appl Mech Eng 372:113377
Li HY, Kafka OL, Gao JY, Yu C, Nie YH, Zhang L, Tajdari M, Tang S, Guo X, Li G, Tang SQ, Cheng GD, Liu WK (2019) Clustering discretization methods for generation of material performance databases in machine learning and design optimization. Comput Mech 64(2):281–305
Kollmann HT, Abueidda DW, Koric S, Guleryuz E, Sobh NA (2020) Deep learning for topology optimization of 2D metamaterials. Mater Des 196:109098
Fernández M, Fritzen F, Weeger O (2021) Material modeling for parametric, anisotropic finite strain hyperelasticity based on machine learning with application in optimization of metamaterials. Int J Numer Meth Eng 123(2):577–609
Chen JL, Wang K, Zhang C, Wang B (2018) An efficient statistical approach to design 3D-printed metamaterials for mimicking mechanical properties of soft biological tissues. Addit Manuf 24:341–352
Zhu L, Sun L, Wang XY, Li N (2021) Optimisation of three-dimensional hierarchical structures with tailored lattice metamaterial anisotropy. Mater Design 210
Wu RT, Liu TW, Jahanshahi MR, Semperlotti F (2021) Design of one-dimensional acoustic metamaterials using machine learning and cell concatenation. Struct Multidiscip Optim 63(5):2399–2423
Wilt JK, Yang C, Gu GX (2020) Accelerating auxetic metamaterial design with deep learning. Adv Eng Mater. https://doi.org/10.1002/adem.202070018
Wang LW, Yerramilli S, Iyer A, Apley D, Zhu P, Chen W (2022) Scalable Gaussian processes for data-driven design using big data with categorical factors. J Mech Design. https://doi.org/10.1115/1.4052221
Ma W, Cheng F, Xu Y, Wen Q, Liu Y (2019) Probabilistic representation and inverse design of metamaterials based on a deep generative model with semi-supervised learning strategy. Adv Mater 31(35):e1901111
Ma W, Cheng F, Liu Y (2018) Deep-learning-enabled on-demand design of chiral metamaterials. ACS Nano 12(6):6326–6334
Kuś GI, van der Zwaag S, Bessa MA (2021) Sparse quantum Gaussian processes to counter the curse of dimensionality. Quantum Mach Intell 3(1)
Chang YF, Wang H, Dong QX (2022) Machine learning-based inverse design of auxetic metamaterial with zero Poisson’s ratio. Mater Today Commun 30
Bostanabad R, Chan YC, Wang LW, Zhu P, Chen W (2019) Globally approximate Gaussian processes for big data with application to data-driven metamaterials design. J Mech Design 141(11)
Bonfanti S, Guerra R, Font-Clos F, Rayneau-Kirkhope D, Zapperi S (2020) Automatic design of mechanical metamaterial actuators. Nat Commun 11(1):4162
Bessa MA, Glowacki P, Houlder M (2019) Bayesian machine learning in metamaterial design: fragile becomes supercompressible. Adv Mater 31(48):e1904845
Lee DK, Chan YC, Chen W, Wang LW, van Beek A, Chen W (2023) t-METASET: task-aware acquisition of metamaterial datasets through diversity-based active learning. J Mech Des 145(3):031704
Abu-Mualla M, Huang JD (2023) Inverse design of 3D cellular materials with physics-guided machine learning. Mater Des 232:112103
Sarkar S, Ji AQ, Jermain Z, Lipton R, Brongersma M, Dayal K, Noh HY (2023) Physics-informed machine learning for inverse design of optical metamaterials. Adv Photon Res:2300158
Jain P, Chhabra H, Chauhan U, Singh DK, Anwer TMK, Ahammad SH, Hossain MA, Rashed ANZ (2023) Multiband metamaterial absorber with absorption prediction by assisted machine learning. Mater Chem Phys 307:128180
Brown NK, Garland AP, Fadel GM, Li G (2023) Deep reinforcement learning for the rapid on-demand design of mechanical metamaterials with targeted nonlinear deformation responses. Eng Appl Artif Intell 126:106998
Chacón JM, Caminero MA, García-Plaza E, Núñez PJ (2017) Additive manufacturing of PLA structures using fused deposition modelling: effect of process parameters on mechanical properties and their optimal selection. Mater Des 124:143–157
Shim DS (2021) Effects of process parameters on additive manufacturing of aluminum porous materials and their optimization using response surface method. J Mater Res Technol 15:119–134
Gan ZT, Li HY, Wolff SJ, Bennett JL, Hyatt G, Wagner GJ, Cao J, Liu WK (2019) Data-driven microstructure and microhardness design in additive manufacturing using a self-organizing map. Engineering 5(4):730–735
Yao XL, Moon SK, Bi GJ (2017) A hybrid machine learning approach for additive manufacturing design feature recommendation. Rapid Prototyp J 23(6):983–997
Adnan M, Lu Y, Jones A, Cheng FT, Yeung H (2020) A new architectural approach to monitoring and controlling AM processes. Appl Sci 10(18):6616
Liu R, Liu S, Zhang XL (2021) A physics-informed machine learning model for porosity analysis in laser powder bed fusion additive manufacturing. Int J Adv Manuf Technol 113(7–8):1943–1958
Baturynska I, Martinsen K (2021) Prediction of geometry deviations in additive manufactured parts: comparison of linear regression with machine learning algorithms. J Intell Manuf 32(1):179–200
Shi J, Wu B, Song B, Song J, Li S, Trau D, Lu WF (2018) Learning-based cell injection control for precise drop-on-demand cell printing. Ann Biomed Eng 46(9):1267–1279
Bone JM, Childs CM, Menon A, Poczos B, Feinberg AW, LeDuc PR, Washburn NR (2020) Hierarchical machine learning for high-fidelity 3D printed biopolymers. ACS Biomater Sci Eng 6(12):7021–7031
Ng WL, Chan A, Ong YS, Chua CK (2020) Deep learning for fabrication and maturation of 3D bioprinted tissues and organs. Virtual and Physical Prototyping 15(3):340–358
Xu HQ, Liu QY, Casillas J, Mcanally M, Mubtasim N, Gollahon LS, Wu DZ, Xu CX (2022) Prediction of cell viability in dynamic optical projection stereolithography-based bioprinting using machine learning. J Intell Manuf 33(4):995–1005
Fu Z, Angeline V, Sun W (2021) Evaluation of printing parameters on 3D extrusion printing of pluronic hydrogels and machine learning guided parameter recommendation. Int J Bioprint 7(4):434
Reina-Romo E, Mandal S, Amorim P, Bloemen V, Ferraris E, Geris L (2021) Towards the experimentally-informed in silico nozzle design optimization for extrusion-based bioprinting of shear-thinning hydrogels. Front Bioeng Biotechnol 9:701778
Ruberu K, Senadeera M, Rana S, Gupta S, Chung J, Yue ZL, Venkatesh S, Wallace G (2021) Coupling machine learning with 3D bioprinting to fast track optimisation of extrusion printing. Appl Mater Today 22
Tian S, Stevens R, McInnes BT, Lewinski NA (2021) Machine assisted experimentation of extrusion-based bioprinting systems. Micromachines 12(7):780
Guan J, You S, Xiang Y, Schimelman J, Alido J, Ma X, Tang M, Chen S (2021) Compensating the cell-induced light scattering effect in light-based bioprinting using deep learning. Biofabrication 14(1):015011
Shen X, Yao J, Wang Y, Yang J (2004) Density prediction of selective laser sintering parts based on artificial neural network. International symposium on neural networks. Springer, pp 832–840
Munguía J, Ciurana J, Riba C (2009) Neural-network-based model for build-time estimation in selective laser sintering. Proc Instit Mech Eng Part B 223(8):995–1003
Rong-Ji W, Xin-hua L, Qing-ding W, Lingling W (2008) Optimizing process parameters for selective laser sintering based on neural network and genetic algorithm. Int J Adv Manuf Technol 42(11–12):1035–1042
Wang R-J, Li J, Wang F, Li X, Wu Q (2009) ANN model for the prediction of density in selective laser sintering. Int J Manuf Res 4(3):362–373
Lee SH, Park WS, Cho HS, Zhang W, Leu MC (2001) A neural network approach to the modelling and analysis of stereolithography processes. Proc Instit Mech Eng Part B 215(12):1719–1733
Khadilkar A, Wang J, Rai R (2019) Deep learning-based stress prediction for bottom-up SLA 3D printing process. Int J Adv Manuf Technol 102(5–8):2555–2569
Mohamed OA, Masood SH, Bhowmik JL (2017) Influence of processing parameters on creep and recovery behavior of FDM manufactured part using definitive screening design and ANN. Rapid Prototyp J 23(6):998–1010
Sood AK, Ohdar RK, Mahapatra SS (2012) Experimental investigation and empirical modelling of FDM process for compressive strength improvement. J Adv Res 3(1):81–90
Sood AK, Equbal A, Toppo V, Ohdar RK, Mahapatra SS (2012) An investigation on sliding wear of FDM built parts. CIRP J Manuf Sci Technol 5(1):48–54
Equbal A, Sood AK, Mahapatra S (2011) Prediction of dimensional accuracy in fused deposition modelling: a fuzzy logic approach. Int J Productivity Qual Manag 7(1):22–43
Gorguluarslan RM, Grandhi RV, Choi HJ, Choi SK (2019) Prediction assessment and validation of multiscale models for additively manufactured lattice structures under uncertainty. J Mech Sci Technol 33(3):1365–1379
Vosniakos GC, Maroulis T, Pantelis D (2007) A method for optimizing process parameters in layer-based rapid prototyping. Proc Instit Mech Eng Part B 221(8):1329–1340
Li ZX, Zhang ZY, Shi JC, Wu DZ (2019) Prediction of surface roughness in extrusion-based additive manufacturing with machine learning. Robot Comput-Integr Manuf 57:488–495
Li XY, Zhang MN, Zhou MX, Wang J, Zhu WX, Wu C, Zhang X (2023) Qualify assessment for extrusion-based additive manufacturing with 3D scan and machine learning. J Manuf Process 90:274–285
Mishra A, Jatti VS (2023) Novel coupled genetic algorithm-machine learning approach for predicting surface roughness in fused deposition modeling of polylactic acid specimens. J Mater Eng Perform. https://doi.org/10.1007/s11665-023-08379-2
Veeman D, Sudharsan S, Surendhar GJ, Shanmugam R, Guo L (2023) Machine learning model for predicting the hardness of additively manufactured acrylonitrile butadiene styrene. Mater Today Commun 35:106147
Lu ZL, Li DC, Lu BH, Zhang AF, Zhu GX, Pi G (2010) The prediction of the building precision in the laser engineered net shaping process using advanced networks. Opt Lasers Eng 48(5):519–525
Xiong J, Zhang G, Hu J, Wu L (2012) Bead geometry prediction for robotic GMAW-based rapid manufacturing through a neural network and a second-order regression analysis. J Intell Manuf 25(1):157–163
Herriott C, Spear AD (2020) Predicting microstructure-dependent mechanical properties in additively manufactured metals with machine- and deep-learning methods. Comput Mater Sci 175:109599
Caiazzo F, Caggiano A (2018) Laser direct metal deposition of 2024 Al alloy: trace geometry prediction via machine learning. Materials 11(3):444
Mozaffar M, Paul A, Al-Bahrani R, Wolff S, Choudhary A, Agrawal A, Ehmann K, Cao J (2018) Data-driven prediction of the high-dimensional thermal history in directed energy deposition processes via recurrent neural networks. Manuf Lett 18:35–39
Kumar P, Jain NK (2022) Surface roughness prediction in micro-plasma transferred arc metal additive manufacturing process using K-nearest neighbors algorithm. Int J Adv Manuf Technol 119(5–6):2985–2997
Zhang M, Sun CN, Zhang X, Goh PC, Wei J, Hardacre D, Li H (2019) High cycle fatigue life prediction of laser additive manufactured stainless steel: a machine learning approach. Int J Fatigue 128:105194
Tapia G, Khairallah S, Matthews M, King WE, Elwany A (2017) Gaussian process-based surrogate modeling framework for process planning in laser powder-bed fusion additive manufacturing of 316L stainless steel. Int J Adv Manuf Technol 94(9–12):3591–3603
Zhan ZX, Li H (2021) A novel approach based on the elastoplastic fatigue damage and machine learning models for life prediction of aerospace alloy parts fabricated by additive manufacturing. Int J Fatigue 145
Minkowitz L, Arneitz S, Effertz PS, Amancio ST (2023) Laser-powder bed fusion process optimisation of AlSi10Mg using extra trees regression. Mater Des 227:111718
Tapia G, Elwany AH, Sang H (2016) Prediction of porosity in metal-based additive manufacturing using spatial Gaussian process models. Addit Manuf 12:282–290
Zhan ZX, Li H (2021) Machine learning based fatigue life prediction with effects of additive manufacturing process parameters for printed SS 316L. Int J Fatigue 142
Tridello A, Ciampaglia A, Berto F, Paolino DS (2023) Assessment of the critical defect in additive manufacturing components through machine learning algorithms. Appl Sci 13(7):4294
Centola A, Ciampaglia A, Tridello A, Paolino DS (2023) Machine learning methods to predict the fatigue life of selectively laser melted Ti6Al4V components. Fatigue Fract Eng Mater Struct 46(11):4350–4370
Zhang W, Mehta A, Desai PS, Higgs CF III (2017) Machine learning enabled powder spreading process map for metal additive manufacturing (AM). In: 2017 International solid freeform fabrication symposium. University of Texas at Austin
Chen H, Zhao YF (2015) Learning algorithm based modeling and process parameters recommendation system for binder jetting additive manufacturing process. In: International design engineering technical conferences and computers and information in engineering conference. American Society of Mechanical Engineersp. V01AT02A029
Xia CY, Pan ZX, Polden J, Li HJ, Xu YL, Chen SB (2022) Modelling and prediction of surface roughness in wire arc additive manufacturing using machine learning. J Intell Manuf 33(5):1467–1482
Yaseer A, Chen HP (2021) Machine learning based layer roughness modeling in robotic additive manufacturing. J Manuf Process 70:543–552
Xiao X, Waddell C, Hamilton C, Xiao H (2022) Quality prediction and control in wire arc additive manufacturing via novel machine learning framework. Micromachines 13(1):137
Shi J, Song JC, Song B, Lu WF (2019) Multi-objective optimization design through machine learning for drop-on-demand bioprinting. Engineering 5(3):586–593
Fu YZ, Downey A, Yuan L, Pratt A, Balogun Y (2021) In situ monitoring for fused filament fabrication process: a review. Addit Manuf 38:101749
Lukin S, Uzarevic K, Halasz I (2021) Raman spectroscopy for real-time and in situ monitoring of mechanochemical milling reactions. Nat Protoc 16(7):3492–3521
Usha S (2021) In situ monitoring of metal additive manufacturing process: a review. additive manufacturing. Elsevier, pp 275–299
Mahato V, Obeidi MA, Brabazon D, Cunningham P (2020) An evaluation of classification methods for 3D printing time-series data. IFAC Papersonline 53(2):8211–8216
Mahato V, Obeidi MA, Brabazon D, Cunningham P (2022) Detecting voids in 3D printing using melt pool time series data. J Intell Manuf 33(3):845–852
Guo QL, Zhao C, Escano LI, Young Z, Xiong LH, Fezzaa K, Everhart W, Brown B, Sun T, Chen LY (2018) Transient dynamics of powder spattering in laser powder bed fusion additive manufacturing process revealed by in situ high-speed high-energy X-ray imaging. Acta Mater 151:169–180
Leung CLA, Marussi S, Atwood RC, Towrie M, Withers PJ, Lee PD (2018) In situ X-ray imaging of defect and molten pool dynamics in laser additive manufacturing. Nat Commun 9(1):1355
Wang TJ, Kwok TH, Zhou C, Vader S (2018) droplet inspection and closed-loop control system using machine learning for liquid metal jet printing. J Manuf Syst 47:83–92
Zhang YJ, Hong GS, Ye DS, Zhu KP, Fuh JYH (2018) Extraction and evaluation of melt pool, plume and spatter information for powder-bed fusion AM process monitoring. Mater Des 156:458–469
Wu D, Wei Y, Terpenny J (2018) Surface roughness prediction in additive manufacturing using machine learning. In: International manufacturing science and engineering conference. American Society of Mechanical Engineers, p. V003T02A018
Syam WP, Leach R, Rybalcenko K, Gaio A, Crabtree J (2018) In-process measurement of the surface quality for a novel finishing process for polymer additive manufacturing. 15th Cirp Conf Comput Aid Tolerancing Cirp Cat 2018 75:108–113
Halsey W, Rose D, Scime L, Dehoff R, Paquit V (2021) Localized defect detection from spatially mapped, process data with machine learning. Front Mech Eng. https://doi.org/10.3389/fmech.2021.767444
Rossi A, Moretti M, Senin N (2021) Layer inspection via digital imaging and machine learning for in-process monitoring of fused filament fabrication. J Manuf Process 70:438–451
Wu HX, Yu ZH, Wang Y (2019) Experimental study of the process failure diagnosis in additive manufacturing based on acoustic emission. Measurement 136:445–453
Liu J, Hu YM, Wu B, Wang Y (2018) An improved fault diagnosis approach for FDM process with acoustic emission. J Manuf Process 35:570–579
Scheffel RM, Fröhlich AA, Silvestri M (2021) Automated fault detection for additive manufacturing using vibration sensors. Int J Comput Integr Manuf 34(5):500–514
Delli U, Chang S (2018) Automated process monitoring in 3D printing using supervised machine learning. In: 46th SME North American Manufacturing Research Conference, NAMRC 46 26: 865–870.
Chen LQ, Yao XL, Xu P, Moon SK, Bi GJ (2021) Rapid surface defect identification for additive manufacturing with in situ point cloud processing and machine learning. Virtual Phys Prototyp 16(1):50–67
Ren WJ, Wen GR, Zhang ZF, Mazumder J (2022) Quality monitoring in additive manufacturing using emission spectroscopy and unsupervised deep learning. Mater Manuf Processes 37(11):1339–1346
Hossain MS, Taheri H (2021) In situ process monitoring for metal additive manufacturing through acoustic techniques using wavelet and convolutional neural network (CNN). Int J Adv Manuf Technol 116(11–12):3473–3488
Liu M, Senin N, Leach R, Lehmann P, Osten W, Albertazzi Gonçalves A (2021) Intelligent quality monitoring for additive manufactured surfaces by machine learning and light scattering. Optic Measure Syst Ind Inspection XII
Yuan BD, Guss GM, Wilson AC, Hau-Riege SP, DePond PJ, McMains, SM, Matthews J, Giera B (2018) Machine-learning-based monitoring of laser powder bed fusion. Adv Mater Technol 3(12)
Bugatti M, Colosimo BM (2022) Towards real-time in situ monitoring of hot-spot defects in L-PBF: a new classification-based method for fast video-imaging data analysis. J Intell Manuf 33(1):293–309
Sun WB, Zhang ZH, Ren WJ, Mazumder J, Jin JH (2022) In situ monitoring of optical emission spectra for microscopic pores in metal additive manufacturing. J Manuf Sci Eng—Trans ASME 144(1)
Montazeri M, Nassar AR, Dunbar AJ, Rao P (2020) In-process monitoring of porosity in additive manufacturing using optical emission spectroscopy. IISE Tran 52(5):500–515
Scime L, Beuth J (2019) Using machine learning to identify in situ melt pool signatures indicative of flaw formation in a laser powder bed fusion additive manufacturing process. Addit Manuf 25:151–165
Scime L, Beuth J (2018) A multi-scale convolutional neural network for autonomous anomaly detection and classification in a laser powder bed fusion additive manufacturing process. Addit Manuf 24:273–286
Scime L, Beuth J (2018) Anomaly detection and classification in a laser powder bed additive manufacturing process using a trained computer vision algorithm. Addit Manuf 19:114–126
Larsen S, Hooper PA (2022) Deep semi-supervised learning of dynamics for anomaly detection in laser powder bed fusion. J Intell Manuf 33(2):457–471
Nguyen NV, Hum AJW, Do T, Tran T (2023) Semi-supervised machine learning of optical monitoring data for anomaly detection in laser powder bed fusion. Virtual Phys Prototyp 18(1):e2129396
Wasmer K, Le-Quang T, Meylan B, Shevchik SA (2018) In situ quality monitoring in AM using acoustic emission: a reinforcement learning approach. J Mater Eng Perform 28(2):666–672
Shevchik SA, Masinelli G, Kenel C, Leinenbach C, Wasmer K (2019) Deep learning for and real-time quality monitoring in additive manufacturing using acoustic emission. IEEE Trans Ind Inf 15(9):5194–5203
Shevchik SA, Kenel C, Leinenbach C, Wasmer K (2018) Acoustic emission for in situ quality monitoring in additive manufacturing using spectral convolutional neural networks. Addit Manuf 21:598–604
Gobert C, Reutzel EW, Petrich J, Nassar AR, Phoha S (2018) Application of supervised machine learning for defect detection during metallic powder bed fusion additive manufacturing using high resolution imaging. Addit Manuf 21:517–528
Snow Z, Diehl B, Reutzel EW, Nassar A (2021) Toward in situ flaw detection in laser powder bed fusion additive manufacturing through layerwise imagery and machine learning. J Manuf Syst 59:12–26
Petrich J, Snow Z, Corbin D, Reutzel EW (2021) Multi-modal sensor fusion with machine learning for data-driven process monitoring for additive manufacturing. Addit Manuf 48:102364
Ghayoomi Mohammadi M, Mahmoud D, Elbestawi M (2021) On the application of machine learning for defect detection in L-PBF additive manufacturing. Opt Laser Technol 143:107338
Kononenko DY, Nikonova V, Seleznev M, van den Brink J, Chernyavsky D (2023) An in situ crack detection approach in additive manufacturing based on acoustic emission and machine learning. Addit Manuf Lett 5:100130
Yadav P, Singh VK, Joffre T, Rigo O, Arvieu C, Le Guen E, Lacoste E (2020) Inline drift detection using monitoring systems and machine learning in selective laser melting. Adv Eng Mater. https://doi.org/10.1002/adem.202000660
Li JC, Zhou Q, Huang XF, Li ML, Cao LC (2023) In situ quality inspection with layer-wise visual images based on deep transfer learning during selective laser melting. J Intell Manuf 34(2):853–867
Li J, Cao L, Xu J, Wang S, Zhou Q (2022) In situ porosity intelligent classification of selective laser melting based on coaxial monitoring and image processing. Measurement 187:110232
Westphal E, Seitz H (2021) A machine learning method for defect detection and visualization in selective laser sintering based on convolutional neural networks. Addit Manuf 41:101965
Entekhabi E, Haghbin Nazarpak M, Sedighi M, Kazemzadeh A (2020) Predicting degradation rate of genipin cross-linked gelatin scaffolds with machine learning. Mater Sci Eng C Mater Biol Appl 107:110362
Bermejillo Barrera MD, Franco-Martinez F, Diaz Lantada A (2021) Artificial intelligence aided design of tissue engineering scaffolds employing virtual tomography and 3d convolutional neural networks. Materials 14(18):5278
Kondiah PJ, Kondiah PPD, Choonara YE, Marimuthu T, Pillay V (2020) A 3D bioprinted pseudo-bone drug delivery scaffold for bone tissue engineering. Pharmaceutics 12(2):166
Conev A, Litsa EE, Perez MR, Diba M, Mikos AG, Kavraki LE (2020) Machine learning-guided three-dimensional printing of tissue engineering scaffolds. Tissue Eng Part A 26(23–24):1359–1368
Vatankhah E, Semnani D, Prabhakaran MP, Tadayon M, Razavi S, Ramakrishna S (2014) Artificial neural network for modeling the elastic modulus of electrospun polycaprolactone/gelatin scaffolds. Acta Biomater 10(2):709–721
Asadi-Eydivand M, Solati-Hashjin M, Fathi A, Padashi M, AbuOsman NA (2016) Optimal design of a 3D-printed scaffold using intelligent evolutionary algorithms. Appl Soft Comput 39:36–47
Rabiee SM, Mozaffari A, Fathi A (2016) Investigation of hydroxyapatite dicalcium phosphate scaffold properties using a Lamarckian immune neural network. Int J Comput Appl Technol 53(4):323–335
Sujeeun LY, Goonoo N, Ramphul H, Chummun I, Gimie F, Baichoo S, Bhaw-Luximon A (2020) Correlating in vitro performance with physico-chemical characteristics of nanofibrous scaffolds for skin tissue engineering using supervised machine learning algorithms. R Soc Open Sci 7(12):201293
Liu W, Zhang Y, Lyu Y, Bosiakov S, Liu Y (2023) Inverse design of anisotropic bone scaffold based on machine learning and regenerative genetic algorithm. Front Bioeng Biotechnol 11:1241151
Cilla M, Borgiani E, Martinez J, Duda GN, Checa S (2017) Machine learning techniques for the optimization of joint replacements: application to a short-stem hip implant. PLoS ONE 12(9):e0183755
Borjali A, Monson K, Raeymaekers B (2019) Predicting the polyethylene wear rate in pin-on-disc experiments in the context of prosthetic hip implants: deriving a data-driven model using machine learning methods. Tribol Int 133:101–110
Chanda S, Gupta S, Pratihar DK (2015) Effects of interfacial conditions on shape optimization of cementless hip stem: an investigation based on a hybrid framework. Struct Multidiscip Optim 53(5):1143–1155
Chanda S, Gupta S, Pratihar DK (2016) A combined neural network and genetic algorithm based approach for optimally designed femoral implant having improved primary stability. Appl Soft Comput 38:296–307
Kang YJ, Yoo JI, Cha YH, Park CH, Kim JT (2020) Machine learning-based identification of hip arthroplasty designs. J Orthop Translat 21:13–17
Borjali A, Chen AF, Muratoglu OK, Morid MA, Varadarajan KM (2020) Detecting total hip replacement prosthesis design on plain radiographs using deep convolutional neural network. J Orthop Res 38(7):1465–1471
Milimonfared R, Oskouei RH, Taylor M, Solomon LB (2018) An intelligent system for image-based rating of corrosion severity at stem taper of retrieved hip replacement implants. Med Eng Phys 61:13–24
Akkad K, Mehboob H, Alyamani R, Tarlochan F (2023) A machine-learning-based approach for predicting mechanical performance of semi-porous hip stems. J Funct Biomater 14(3):156
Alkentar R, Mankovits T (2023) Optimization of additively manufactured and lattice-structured hip implants using the linear regression algorithm from the scikit-learn library. Crystals 13(10):1513
Thomas KA, Dey S, Sultana N, Sarkar K, Datta S (2020) Design of Ti composite with bioactive surface for dental implant. Mater Manuf Processes 35(6):643–651
Sadat R, Khalili M, Nazari M (2016) A hybrid method to predict success of dental implants. Int J Adv Comput Sci App 7(5)
Lee DW, Kim SY, Jeong SN, Lee JH (2021) Artificial intelligence in fractured dental implant detection and classification: evaluation using dataset from two dental hospitals. Diagnostics 11(2):233
Cha JY, Yoon HI, Yeo IS, Huh KH, Han JS (2021) Peri-implant bone loss measurement using a region-based convolutional neural network on dental periapical radiographs. J Clin Med 10(5):1009
Zhang H, Shan J, Zhang P, Chen X, Jiang H (2020) Trabeculae microstructure parameters serve as effective predictors for marginal bone loss of dental implant in the mandible. Sci Rep 10(1):18437
Maudes J, Bustillo A, Guerra AJ, Ciurana J (2017) Random Forest ensemble prediction of stent dimensions in microfabrication processes. Int J Adv Manuf Technol 91(1–4):879–893
Dong Y, Que L, Jia Q, Xi Y, Zhuang J, Li J, Liu H, Chen W, Huang M (2022) Predicting reintervention after thoracic endovascular aortic repair of Stanford type B aortic dissection using machine learning. Eur Radiol 32(1):355–367
Wang Z, Jenkins MW, Linderman GC, Bezerra HG, Fujino Y, Costa MA, Wilson DL, Rollins AM (2015) 3-D stent detection in intravascular OCT using a Bayesian network and graph search. IEEE Trans Med Imaging 34(7):1549–1561
Sampedro-Gomez J, Dorado-Diaz PI, Vicente-Palacios V, Sanchez-Puente A, Jimenez-Navarro M, SanRoman JA, Galindo-Villardon P, Sanchez PL, Fernandez-Aviles F (2020) Machine learning to predict stent restenosis based on daily demographic, clinical and angiographic characteristics. Can J Cardiol 36(10):1624–1632
Tang CX, Guo BJ, Schoepf JU, Bayer RR 2nd, Liu CY, Qiao HY, Zhou F, Lu GM, Zhou CS, Zhang LJ (2021) Feasibility and prognostic role of machine learning-based FFR(CT) in patients with stent implantation. Eur Radiol 31(9):6592–6604
Wang Y, Zhu K, Li Y, Lv Q, Fu G, Zhang W (2020) A machine learning-based approach for the prediction of periprocedural myocardial infarction by using routine data. Cardiovasc Diagn Ther 10(5):1313–1324
Sakai T, Li H, Shimada T, Kita S, Iida M, Lee C, Nakano T, Yamaguchi S, Imazato S (2023) Development of artificial intelligence model for supporting implant drilling protocol decision making. J Prosthodont Res 67(3):360–365
Rekawek P, Herbst EA, Suri A, Ford BP, Rajapakse CS, Panchal N (2023) Machine learning and artificial intelligence: a web-based implant failure and peri-implantitis prediction model for clinicians. Int J Oral Maxillofac Implants 38(3):576–582b
Mohammad-Rahimi H, Motamedian SR, Pirayesh Z, Haiat A, Zahedrozegar S, Mahmoudinia E, Rohban MH, Krois J, Lee JH, Schwendicke F (2022) Deep learning in periodontology and oral implantology: a scoping review. J Periodontal Res 57(5):942–951
Lim H-K, Kwon Y-J, Lee E-S (2020) Application of artificial intelligence in the identification of dental implant systems: a literature review. J Dent Implant Res 39(4):48–52
Hofmann P, Kunz A, Schmidt F, Beuer F, Duddeck D (2023) Influence of exposure of customized dental implant abutments to different cleaning procedures: an in vitro study using AI-assisted SEM/EDS analysis. Int J Implant Dent 9(1):33
Elgarba BM, Van Aelst S, Swaity A, Morgan N, Shujaat S, Jacobs R (2023) Deep learning-based segmentation of dental implants on cone-beam computed tomography images: a validation study. J Dent 137:104639
Benakatti VB, Nayakar RP, Anandhalli M (2021) Machine learning for identification of dental implant systems based on shape—a descriptive study. J Indian Prosthodont Soc 21(4):405–411
Ramachandran RA, Barao VAR, Ozevin D, Sukotjo C, Srinivasa PP, Mathew M (2023) Early predicting tribocorrosion rate of dental implant titanium materials using random forest machine learning models. Tribol Int 187:108735
Alqutaibi AY (2023) Artificial intelligence models show potential in recognizing the dental implant type, predicting implant success, and optimizing implant design. J Evid Based Dent Pract 23(1):101836
Alharbi MT, Almutiq MM (2022) Prediction of dental implants using machine learning algorithms. J Healthc Eng 2022:7307675
Liu CH, Lin CJ, Hu YH, You ZH (2018) Predicting the failure of dental implants using supervised learning techniques. Appl Sci 8(5):698
Ha SR, Park HS, Kim EH, Kim HK, Yang JY, Heo J, Yeo IL (2018) A pilot study using machine learning methods about factors influencing prognosis of dental implants. J Adv Prosthodont 10(6):395–400
Li JJ, Dunstan CR, Entezari A, Li Q, Steck R, Saifzadeh S, Sadeghpour A, Field JR, Akey A, Vielreicher M (2019) A novel bone substitute with high bioactivity, strength, and porosity for repairing large and load-bearing bone defects. Adv Healthcare Mater 8(8):1801298
Acknowledgements
We acknowledge financial supports from the Australian Research Council through the Discovery (DP230103180) and Linkage (LP180101352) schemes.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work is supported by the Australian Research Council through the Discovery project (Grant No. DP230103180) and Linkage project (Grant No. LP180101352).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, C., Xu, Y., Fang, J. et al. Machine Learning in Biomaterials, Biomechanics/Mechanobiology, and Biofabrication: State of the Art and Perspective. Arch Computat Methods Eng (2024). https://doi.org/10.1007/s11831-024-10100-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11831-024-10100-y