Abstract
Taxodium is a genus renowned for its fast growth, good form and tolerance of flooding, salt, alkalinity, disease and strong winds. In this study, a genetic linkage map was constructed using sequence-related amplified polymorphism (SRAP) and simple sequence repeat (SSR) markers based on an F1 population containing 148 individuals generated from a cross between T. ‘Zhongshanshan 302’ and T. mucronatum. The map has a total length of 976.5 cM, with a mean distance of 7.0 cM between markers, and contains 34 linkage groups with 179 markers (171 SRAPs and 8 SSRs). Quantitative trait loci (QTLs) affecting growth traits, such as seedling height, basal diameter and crown width, were detected based on the constructed linkage map. Four significant QTLs were identified, three of which, namely qtSH-1 for seedling height, qtBD-1 for basal diameter and qtCW-1 for crown width, were located at 2.659 cM of LG7 with logarithm odds values of 3.72, 3.49 and 3.93, respectively, and explained 24.9, 27.0 and 21.7 % of the total variation of the three grown traits, respectively. Another QTL for crown width (qtCW-2) was detected at 1.0 cM on LG13, with a logarithm of odds value of 3.15, and explained 31.7 % of the total variation of crown width. This is the first report on the construction of a genetic linkage map and QTL analysis in Taxodium, laying the groundwork for the construction of a high-density genetic map and QTL mapping in the genus Taxodium.
Similar content being viewed by others
Background
Taxodium is a genus containing three coniferous species, viz. Taxodium distichum, Taxodium ascendens and Taxodium mucronatum (Qi et al. 2014), which are allogamous, wind-pollinated, and diploid with a haploid chromosome number (n) of 11 (2n = 22). T. distichum is native to the southeastern United States, from Delaware to Texas, and inland up the Mississippi River to southern Indiana. It is highly resistant to Cercosporidium needle blight and tolerant of flooding, salt, alkalinity and strong winds (Creech et al. 2011). T. mucronatum is native to Mexico, much of Guatemala, the tip of South Texas and New Mexico. It is more tolerant of salt and alkaline soils, but less tolerant of flooding and cercosporidium needle blight (Creech et al. 2011; Zhou et al. 2010). T. ‘Zhongshanshan 302’ is a superior clone selected from a controlled cross between T. distichum and T. mucronatum (Wang et al. 2015). It is well known for its fast growth, good form, and strong adaptability to a wide range of soils and climates (Cheng et al. 2015). It is also relatively pest-free and has a higher tolerance of salt, alkalinity and flooding (Zhou et al. 2010; Qi et al. 2014). Thus, it has great ecological and economic potential (Cheng et al. 2015). T. ‘Zhongshanshan 302’ is registered as a Chinese national variety (Zhou et al. 2010; Wang et al. 2015) and has been widely planted in urban areas and wetlands of eastern China (Zhou et al. 2010; Wang et al. 2015; Cheng et al. 2015; Qi et al. 2014).
Linkage maps facilitate not only gene tagging, map-based cloning (Muchero et al. 2015; Yang et al. 2013a, b), comparative genomic studies (Moriguchi et al. 2012), construction of physical maps, the assembly of whole-genomes (Martínez-García et al. 2013; Marone et al. 2012), and understanding of genome structure and evolution (Scotti et al. 2005; Jermstad et al. 2011), but also molecular marker-assisted selection (MAS) (Moriguchi et al. 2012; Jena et al. 2006) and quantitative trait loci (QTLs) mapping (Moriguchi et al. 2012). In recent decades, genetic maps have been constructed for many conifers, such as Picea abies (Scotti et al. 2005; Acheré et al. 2004), Picea mariana (Kang et al. 2010; Pelgas et al. 2005), Pinus taeda (Martínez-García et al. 2013; Eckert et al. 2009; Temesgen et al. 2001), Pinus radiata (Moraga-Suazo et al. 2014), Pinus pinaster (Lepoittevin et al. 2012; Rittera et al. 2002), Pinus koraiensis (Chen et al. 2010a, b), Pinus elliottii (Yang et al. 2013a; Shepherd et al. 2003) and Pinus lambertiana (Jermstad et al. 2011). Additionally, MAS for QTLs has been reported in nearly all crop species (Moriguchi et al. 2012; Jena et al. 2006; Zhong et al. 2006). To the best of our knowledge, the majority of genetic maps in conifers have been reported in Pinaceae. In Taxodiaceae, just Cryptomeria and Cunninghamia have had several unsaturated maps developed, containing few DNA molecular markers. Tong and Shi (2004) constructed two maps for C. lanceolata using 94 and 101 AFLP markers. In 2012, Moriguchi et al. (2012) reported a genetic map for C. japonica, in which 1261 SNP markers were mapped on 11 LGs. To date, it is the only high-density genetic linkage map that has been constructed predominantly in Taxodiaceae.
Compared with traditional tree improvement approaches that involve the selection of superior trees based on growth characteristics, wood properties or biotic and abiotic stress responses during long growth cycles (Nowicka et al. 2013), QTL mapping offers new opportunities for effective tree breeding (Yoshimaru et al. 1998). QTLs associated with phenotypic variability can be identified accurately by molecular markers in a suitable segregating population (Moraga-Suazo et al. 2014), which has important consequences for employing markers in trees at early stages (Lerceteau et al. 2000) and provides huge potential benefits for improving traits that are difficult, expensive and time-consuming to measure (Nowicka et al. 2013). In recent years, QTL studies based on genetic linkage maps have been reported in several tree species (Nowicka et al. 2013; Lerceteau et al. 2000, 2001; Sewell et al. 2002; Pot et al. 2006; Ukrainetz et al. 2008; Pelgas et al. 2011; Wheelerl et al. 2005).
The use of sequence-related amplified polymorphisms (SRAPs) was initially proposed by Li and Quiros (2001). The PCR-based DNA marker system aims to amplify open reading frames with particular primer pairs (Sun et al. 2006; Li and Quiros 2001; Guo et al. 2014), and its polymorphisms originate from the variations in promoter, intron and spacer lengths among individuals and species (Sun et al. 2006; Li and Quiros 2001). SRAP markers have many advantages, including reliability, reproducibility, simplicity, high efficiency, moderate throughput ratios and easy isolation of bands (Yang et al. 2013a; Chen et al. 2010b; Li and Quiros 2001; Guo et al. 2014). Furthermore, it can target functional genes and detect any base changes, insertions or deletions in a sequence (Yang et al. 2013a; Chen et al. 2010b). Therefore, SRAP can be employed in cDNA fingerprinting, genetic map construction, QTL mapping, comparative genetics and genetic diversity assessments (Yang et al. 2013a; Sun et al. 2006). Yu et al. (2009) established and optimized the SRAP-PCR reaction system in Taxodium and successfully identified authentic hybrids generated from a cross of T. distichum and T. mucronatum using 12 polymorphic SRAP combinations, concluding that SRAP markers are useful and efficient molecular markers in Taxodium.
Due to its origin, there are two types of SSR microsatellite markers: genomic SSRs (gSSRs) and expressed sequence tag derived SSRs (EST-SSRs) (Wang et al. 2015; Yang et al. 2013b; Poncet et al. 2006). Compared with gSSRs derived from traditional methods involving the construction of genomic DNA libraries, probe hybridization, cloning and sequencing (Hu et al. 2010; Huang et al. 2011), the development of EST-SSRs with the availability of unaccountable ESTs in public databases, and the advent of Next Generation Sequencing systems, has become a convenient and cost-effective option (Wang et al. 2015; Cheng et al. 2015). SSR markers are considered effective and powerful for assessing genetic diversity and quantifying population genetic structures, relatedness and evolution. This is also true for constructing genetic linkage maps and determining QTLs based on their characteristics of co-dominance, abundance, wide distribution over the genome, and high level of polymorphisms, transferability and reproducibility (Wang et al. 2015; Cheng et al. 2015; Gaudet et al. 2008; Canli 2004; Liu et al. 2014). Thus, microsatellite markers have been widely applied to the construction of genetic linkage maps in forest trees, such as Eucalyptus grandis (García et al. 2011), P. cerasus (Canli 2004), P. nigra (Gaudet et al. 2008), P. koraiensis (Chen et al. 2010b), P. abies (Acheré et al. 2004), P. pinaster (Rittera et al. 2002), P. elliottii and P. caribaea (Yang et al. 2013a).
This paper reports a first genetic linkage map for Taxodium that was constructed using SRAP and SSR markers. Several QTLs associated with seedling height (SH), basal diameter (BD) and crown width (CW) were detected and characterized. The results provide useful information for potential associations between DNA markers and growth traits, and facilitate our understanding of the genome architecture and organization of Taxodium.
Methods
Plant material and DNA extraction
An F1 population comprised of 148 individuals generated from a cross of Taxodium ‘Zhongshanshan 302’ and T. mucronatum was used as the mapping population. This family was grown in a nursery at the Institute of Botany, Chinese Academy of Sciences in Jiangsu Province (32°02′ N, 118°28′ E; elevation 30 m). T. ‘Zhongshanshan 302’ (T. distichum × T. mucronatum) is a superior clone that was selected in 1988 (Zhou et al. 2010). All of the F1 progeny were previously identified as authentic hybrids (Wang et al. 2015). Genomic DNA was extracted from the fresh leaves of each progeny using a modified CTAB method (Wang et al. 2015; Tsumura et al. 1995). The concentration of the extracted DNA was standardized (Moriguchi et al. 2012), and the DNA samples were then stored at –20 °C.
The SH, BD and CW data for each progeny at 4 years of age were measured using diameter tape and Vernier calipers. The location of the BD measurements was on the trunk, 20 cm above the ground. The CW of each seedling was calculated using the mean value of canopy diameter measured along two different orientations: from south to north and from east to west.
Correlation analyses of the SH, BD and CW of the F1 population at 4 years of age were performed by SAS 6.12 statistical software (Yao et al. 2016).
SRAP and EST-SSR profiling
In total, 224 pairs of SRAP primer combinations, created using 14 forward and 16 reverse primers (Table 1; Yang et al. 2013a; Li and Quiros 2001; Yu et al. 2009; Wang et al. 2005), and 503 EST-SSR primer pairs, developed from the transcriptome sequences of T. ‘Zhongshanshan 405’ (Wang et al. 2015; Cheng et al. 2015), were used to screen for gene polymorphisms in the parents and four F1 hybrid individuals. The polymorphic primer combinations were used in PCR for the mapping population. Loci with null alleles were removed from map construction.
SRAP-PCR was performed in a total volume of 10 µL containing 50 ng genomic DNA, 1 µL of 10× PCR buffer, 2.0 mmol L−1 MgCl2, 0.2 mmol L−1 dNTPs, 0.3 µmol L−1 primers, and 0.5 U Tag DNA polymerase (Shanghai Generay Biotech Co. Ltd, Sanghai, China). PCR reactions were performed in a TC-412 PCR thermal cycler (Bibby Scientific, Stone, United Kingdom) under the following thermal conditions: predenaturation at 94 °C for 4 min; followed by five cycles of denaturation at 94 °C for 1 min, annealing at 37 °C for 1 min, and extension at 72 °C for 1 min; then 35 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 7 min.
SSR-PCR amplification was performed in a 10 µL PCR mixture containing 20 ng genomic DNA, 1 µL of 10× PCR buffer, 3.75 mmol L−1 MgCl2, 0.4 mmol L−1 dNTPs, 0.25 µmol L−1 primers, and 0.5 U Tag DNA polymerase. The PCR was performed under the following conditions: an initial predenaturation at 94 °C for 3 min, followed by 30 cycles of 30 s at 94 °C, 45 s at the annealing temperature of 59 °C and 45 s at 72 °C, ending with a final extension at 72 °C for 7 min.
The PCR products were stored at 4 °C before being separated on 12 % non-denaturing polyacrylamide gels. Electrophoresis was conducted in 0.5 × TBE buffer (pH 8.0) at 120 V for 1–1.5 h. A 50-bp DNA ladder marker (Takara Biotechnology Co. Ltd, Dalian, China) was used as the molecular standard.
Genetic linkage map construction
Each band in the electrophoresis gel represented an allelic locus, and the genotypes of individuals from the mapping population could be reconstructed by counting the location and number of bands detected. JoinMap 4.0 was used to construct the linkage map (Van Ooijen 2006). The mapping population in this study could be considered as a cross-pollination population because the genetic background of the two parents was heterozygous. Three segregation type codes <lmxll>, <nnxnp> and <hkxhk> were used to score heterozygous loci in the female parent (T. ‘Zhongshanshan 302’), the male parent (T. mucronatum), and in both parents, respectively. A Chi square (χ2) test was applied to detect whether the inherited alleles of the mapping population were in compliance with the Mendelian segregation ratios. For alleles that were heterozygous in only one of the parents, the segregation ratio across the mapping population was tested against a 1:1 ratio. However, fragments that were heterozygous in both parents were tested against a 3:1 or 1:2:1 ratio. The segregation patterns of markers that did not fit either ratio (P < 0.05) were treated as distorted. Kosambi′s mapping function was used to convert the recombination frequency to a genetic map distance (Kosambi 1944). The “group” command with a logarithm of odds (LOD) threshold of 9.0 and recombination frequency of 0.3 was used to determine all of the linkage groups (LGs). Images of linkage maps were drawn using MapChart 2.1 software (Voorrips 2002).
Estimation of genome length and map coverage
The observed genome length, Go, for the linkage map was calculated as the sum of the sizes of the linkage groups. The expected genome size (Ge) was estimated using Ge = ∑Li[(ki + 1)/(ki − 1)] described by Chakravarti et al. (1991), in which Li is the size of the ith LG (cM) and ki is the number of marker loci on the ith LG. Genome coverage was estimated using the ratio between the observed and the expected genome lengths, i.e. Go/Ge.
QTL analysis
A QTL mapping analysis was performed using interval mapping methods implemented by MapQTL 5.0 (Van Ooijen 2004). A QTL was indicated when the LOD value was higher than the threshold determined using 1000 permutations at a significance level of P = 0.05. The specific location of the QTL was determined using the maximum LOD score in the interval, and confidence intervals (95 %) associated with QTL locations were set as the map intervals corresponding to a 1-point LOD decline on either side of the maximum LOD (Guo et al. 2014). QTLs were named starting with ‘qt’, followed by the abbreviated trait name (SH, BD, or CW) and the number assigned to the QTL.
Female additive (Af), male additive (Am) and dominant (D) effects of the QTLs were estimated using Af = [(uac + uad) − (ubc + ubd)]/4, Am = [(uac + ubc) − (uad + ubd)]/4, and D = [(uac + ubd) − (uad + ubc)]/4, respectively, where, uac, uad, ubc and ubd are the estimated phenotypic means associated with each of the four possible genotypic classes, ac, bc, ad and bd, respectively, derived from a <abxcd> cross (Guo et al. 2014; Leroy et al. 2011; Qin et al. 2008). The detected QTLs were placed on LGs using the MapChart 2.1 software (Voorrips 2002).
Results
Polymorphisms and marker segregation in the mapping population
In total, 113 (50.45 %) of the 224 tested SRAP primer pairs generated 320 polymorphic markers in the F1 population. Of the 320 markers, 209 (65.31 %) and 111 (34.69 %) segregated in 1:1 and 3:1 ratios, respectively. Among the 209 markers with a 1:1 ratio, 150 (71.77 %) originated from the female parent and 59 (28.23 %) originated from the male parent (Table 3). The number of polymorphic markers per primer combination ranged from 1 to 5, with an average of 2.83. Among the 320 SRAP segregation markers, 171 (53.44 %) were mapped to the genetic map, which included 97 maternal markers, 7 paternal markers and 67 parental markers.
Furthermore, 257 (51.09 %) out of 503 EST-SSR primer pairs amplified the expected products, of which 17 (3.38 %; Table 2) amplified polymorphic products in the two parents and 4 F1 individuals. Of the 17 polymorphic markers, 10 (58.82 %) and 7 (41.18 %) segregated in 1:1 and 1:2:1 ratios, respectively. Among the 10 markers with 1:1 ratios, 9 originated from the female parent and 1 originated from the male parent (Table 3). Among the 17 EST-SSR segregation markers, 8 (47.06 %) were mapped to the genetic map.
Among the 337 polymorphic markers (320 SRAPs and 17 EST-SSRs), 284 (84.75 %) showed a normal Mendelian segregation, of which 185 (65.14 %) segregated at 1:1, 82 (28.87 %) segregated at 3:1, and 7 (2.46 %) segregated at 1:2:1 ratios. Additionally, 53 (15.73 %; 50 SRAPs and 3 EST-SSRs) out of the 337 markers showed significant distortions (P < 0.05) from the expected Mendelian segregation ratios (Table 3). Segregation distorted markers were included in the final map only if they did not alter the order of the adjacent markers on the LGs. In total, 49 markers with distorted segregation (47 SRAPs and 2 EST-SSRs) were linked to LGs on the genetic linkage map; 12 and 16 were on LG7 and LG9, respectively, and 21 were on the 15 other LGs (Table 4).
Linkage map construction
The 179 markers mapped were distributed into 18 groups, plus 2 triples and 14 pairs at the 9.0 LOD threshold. The LGs were named LG1 to LG34 based on their lengths. The 34 LGs contained 2–34 markers, and the map size ranged from 3.0 (LG34) to 80 cM (LG1). All of the seven paternal markers (nnxnp) were associated with maternal markers (lmxll) and bi-parental markers (hkxhk) on LG7 and LG9. The map had a total length of 976.5 cM, with a mean distance of 7.0 cM between markers. The gaps between markers ranged from 0.1 to 30.2 cM. The longest gap (30.2 cM) was between TA0106 and TA0440 on LG16, and gaps longer than 20 cM were located on LG2, LG4, LG16, LG19, LG20, LG21 and LG24 (Figs. 1, 2, 3). The expected genome length of Taxodium was 1767.35 cM estimated using method 4 of Chakravarti et al. (1991). The 976.5 cM total size of the linkage map spanned 55.25 % of the estimated Taxodium genome length.
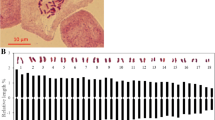
Linkage map from an F1 (T. ‘Zhongshanshan 302’ × T. mucronatum) population, with 171 sequence-related amplified polymorphism (SRAP) and 8 simple sequence repeats (SSRs) markers distributed on 34 linkage groups. The maps had a total length of 976.5 cM. Intervals in cM are shown on the left of each linkage group. The Kosambi function and a logarithm of odds (LOD) threshold of 9.0, and the recombination frequency of 0.3 in JoinMap 4.0 was used to construct the map. MapQTL 5.0 was used to perform quantitative trait locus (QTL) mapping using the interval mapping method. The bars along the linkage maps indicate 1-LOD likelihood intervals for the QTLs. The QTLs are for seedling height (qtSH), basal diameter (qtBD), crown width (qtCW) of mapping population
Linkage map from an F1 (T. ‘Zhongshanshan 302’ × T. mucronatum) population, with 171 sequence-related amplified polymorphism (SRAP) and 8 simple sequence repeats (SSRs) markers distributed on 34 linkage groups. The maps had a total length of 976.5 cM. Intervals in cM are shown on the left of each linkage group. The Kosambi function and a logarithm of odds (LOD) threshold of 9.0, and the recombination frequency of 0.3 in JoinMap 4.0 was used to construct the map. MapQTL 5.0 was used to perform quantitative trait locus (QTL) mapping using the interval mapping method. The bars along the linkage maps indicate 1-LOD likelihood intervals for the QTLs. The QTLs are for seedling height (qtSH), basal diameter (qtBD), crown width (qtCW) of mapping population
Linkage map from an F1 (T. ‘Zhongshanshan 302’ × T. mucronatum) population, with 171 sequence-related amplified polymorphism (SRAP) and 8 simple sequence repeats (SSRs) markers distributed on 34 linkage groups. The maps had a total length of 976.5 cM. Intervals in cM are shown on the left of each linkage group. The Kosambi function and a logarithm of odds (LOD) threshold of 9.0, and the recombination frequency of 0.3 in JoinMap 4.0 was used to construct the map. MapQTL 5.0 was used to perform quantitative trait locus (QTL) mapping using the interval mapping method. The bars along the linkage maps indicate 1-LOD likelihood intervals for the QTLs. The QTLs are for seedling height (qtSH), basal diameter (qtBD), crown width (qtCW) of mapping population
QTL mapping
The results of correlations of the three growth traits of SH, BD and CW showed that there were highly significant (correlations close to 1) among the three traits (Table 5).
Three significant QTLs, including one for SH, qtSH-1, one for BD, qtBD-1, and one for CW, qtCW-1, were detected and allocated to the same position at 2.695 cM on LG7, with LOD values of 3.72, 3.49 and 3.93, respectively, between markers Me14Em8-170 and Me10Em14-200 (1.0 cM from Me14Em8-170 and 2.772 cM from Me10Em14-200). qtSH-1 explained 24.9 % of the total variation of SH, qtBD-1 explained 27.0 % of the total variation of BD and qtCW-1 explained 21.7 % of the total variation of CW. A significant QTL for CW, qtCW-2, was detected on LG13. This QTL explained 31.7 % of the total variation of CW and was located at 1.0 cM on LG13, with a LOD value of 3.15. qtCW-2 was 1.0 cM from Me14Em1-400 and 14.953 cM from Me8Em11-280.
The female and male additive, and the dominant effects of the four QTLs were estimated from an output of the program MapQTL 5.0. The results revealed that both parents had positive effects on the four QTLs, which facilitated the SH, BD and CW of Taxodium. The dominance effects were positive for qtSH-1, qtBD-1 and qtCW-1, but negative for qtCW-2.
The positions, nearest markers and percentages of the phenotypic variance explained by the QTL (PVE), additive and dominant values, and the directions of the four QTLs are shown in Table 6 and Figs. 1, 2, 3.
Discussion
Marker polymorphisms
The genetic map was constructed with 179 markers, of which 150 (46.88 %) of the 320 SRAP markers and 9 (52.94 %) of the 17 EST-SSR markers segregated in the maternal parent (T. ‘Zhongshanshan 302’), 59 (18.44 %) SRAPs and 1 (5.88 %) EST-SSR segregated in the paternal parent (T. mucronatum) and 111 (34.69 %) SRAPs and 7 (41.18 %) EST-SSR segregated in both parents. This difference can be explained by the hybrid origin of T. ‘Zhongshanshan 302’ (Wang et al. 2015).
Previously, SRAP technology was successfully applied to analyze genotypes, authenticate hybrid identifications in Taxodium (Yu et al. 2009) and to assess the genetic diversity and relationships among 18 T. mucronatum individuals of different origins (Zhou et al. 2012). Thus SRAP markers are useful and efficient in Taxodium. According to Guo et al. (2014), Yang et al. (2013a), and Chen et al. (2010a), SRAP technology is an efficient method for constructing genetic maps. In this study, we found that the SRAP markers detected highly polymorphic sites in the Taxodium genome. A total of 320 polymorphic loci were generated by 113 primer combinations, with a mean of 2.83 polymorphic loci per primer combination. The polymorphic locus ratio of SRAP markers was approximately equal to those of P. koraiensis (Chen et al. 2010a) and Zoysiagrass (Guo et al. 2014), but relatively lower than those of other plant species, such as P. elliottii (Yang et al. 2013a), Gossypium hirsutum (Zhang et al. 2009) and Saccharum (Alwala et al. 2008).
Compared with SRAPs, the number of polymorphic SSR markers was insufficient to contribute significantly to the saturation of the map. A total of 17 (3.38 %) SSRs of 503 amplified polymorphic bands in the F1 population revealed polymorphisms. The polymorphic ratio was less than that previously reported in other plants such as 12.24 % of pines (Yang et al. 2013a) and 14.4 % of P. koraiensis (Chen et al. 2010b). However, due to their specificity and co-dominance, SSR markers were very useful for integrating the parental maps (Acheré et al. 2004). In 2014, Wang et al. (2015) detected the cross-family transferability of 60 primer pairs out of 503 EST-SSRs (including 15 polymorphic primer pairs, 15 primer pairs without polymorphisms and 30 primer pairs without products in Taxodium). The results revealed that these primers showed potential cross-family transferability and could be applied to other conifers such as Taxodiaceae, Cupressaceae, Pinaceae and Taxaceae (Wang et al. 2015). Therefore, the potential utility of using these microsatellites in comparison and integration of genetic linkage maps in conifer species will undoubtedly increase in the near future.
Segregation distortion
Segregation distortion is a common phenomenon in the construction of genetic linkage maps (Yang et al. 2013a; Shepherd et al. 2003; Rieseberg et al. 2000) and has been reported in many mapping studies of conifers. Yang et al. (2013a) identified 33.3 and 37.6 % segregation distortion of SRAP, SSR, EST, ISSR markers in P. elliottii var. elliotti and P. caribaea var. hondurensis, respectively, similarly the ratios of skewed AFLP markers were ~30 and 35 %, respectively, in the study of Shepherd et al. (2003). Chen et al. (2010a) detected 25.4 % segregation distortions of SRAP, SSR and ISSR in a P. koraiensis F1 population. Pelgas et al. (2005) detected a 12 % distortion of AFLP, RAPD, SSR and ESTP markers in P. mariana × P. rubens. Iwata et al. (2001) found that 15 (25 %) out of 60 CAPS markers showed a departure from expected segregation ratios in Cryptomeria japonica. He et al. (2000) detected 14.7 % segregation distortion of RAPDs in a Cunninghamia lanceolata F1 population. Such differences in distorted proportions were likely to have been caused by the variance of the population structure, marker types and genetic mechanisms of each species (Shepherd et al. 2003; Guo et al. 2014).
In this study, 53 (15.73 %) out of 337 markers (320 SRAP and 17 SSR) showed significant segregation distortion, which is comparable to results described above. The skewed markers were distributed on 17 LGs, of which 12 and 16 were on LG7 and LG9, respectively, and 21 were on the 15 other LGs. The results were consistent with other reports in which segregation distortion markers were clustered on several linkage groups (Mukai et al. 1995; Guo et al. 2014; Nodari et al. 1993; Kiss et al. 1993). Even though the underlying mechanism for segregation distortion is still debated, it is recognized that this phenomenon might be due to many complicated factors, including environmental factors, experimental errors and biological factors, such as lethal genes, the presence of fragment-complexes, chromosome loss, viability differences among genotypes, gametic and zygotic selection, non-homologous recombination, and the non-homologous or translocation loci on chromosomes (Mukai et al. 1995; Nikaido et al. 2000; Iwata et al. 2001; Cai et al. 2015).
Genetic linkage maps
We present here the first report of a genetic linkage map for Taxodium. The map spanned 976.5 cM, which covered 55.25 % of the estimated genome length, and was assembled using 179 markers, including 171 SRAP and 8 EST-SSR marker loci arranged on 34 LGs. Compared with other species of Taxodiaceae, the map size in our study was shorter than the 1109.1 cM (Iwata et al. 2001), 1405.2 cM (Moriguchi et al. 2012), 1266.1 and 1992.3 cM (Nikaido et al. 2000) of Cryptomeria japonica and the 2282.6 and 2565.8 cM (Tong and Shi 2004) of Cunninghamia lanceolata. It was larger than C. japonica in the study of Mukai et al. (1995), or the 315.3 and 595.2 cM linkage maps of C. lanceolata constructed by He et al. (2000). Furthermore, the number of markers mapped was more than in C. lanceolata using AFLPs (Tong and Shi 2004), RAPDs (He et al. 2000) and C. japonica assembled by CAPS (Nikaido et al. 2000; Iwata et al. 2001), RFLPs, RAPDs (Mukai et al. 1995) and AFLPs (Nikaido et al. 2000). It was less than the map of C. japonica constructed using SNPs (Moriguchi et al. 2012). In this study, 179 markers were randomly distributed among 18 groups, 2 triples and 14 pairs. The lengths of these LGs were quite different, ranging from 3 to 80 cM. The discrepancies in marker numbers and map sizes of different groups have been reported in other species such as P. abies (Scotti et al. 2005), P. taeda (Martínez-García et al. 2013), C. japonica (Nikaido et al. 2000), P. mariana (Kang et al. 2010), P. pinaster (Rittera et al. 2002), P. sylvestris (Yin et al. 2003), P.s nigra (Gaudet et al. 2008), P. cerasus (Canli 2004) and Triticum turgidum (Marone et al. 2012). The average distance between the adjacent markers of this map was 7 cM, and six gaps larger than 20 cM were found on six groups. These large gaps may be associated with the lack of more polymorphic markers and a shortage of marker detection in some regions of chromosome (Cai et al. 2015). The genetic linkage maps constructed will provide a foundation for constructing a high density map for T. ‘Zhongshansa’ in the future.
There are several defects revealed in this map. On the one hand, in view of the huge genome of Taxodium, the map constructed in this paper is only a framework map containing few markers. This defect could be attributed to the closer genetic relationship between the mapping parents and the lower resolution power of marker loci detection means (Wang et al. 2015). Creech et al. (2011), Denny and Arnold (2007) concluded the genus Taxodium is a single species with three botanical varieties: baldcypress (T. distichum var. distichum), pondcypress (T. distichum var. imbricarium), and montezuma cypress (T. distichum var. mexicanum). Therefore, the cross of T. ‘Zhong shanshan302’ and T. mucronatum could be considered as an intraspecific hybridization, and the feeble differences in DNA sequences between the two parents could limit the number of polymorphic markers in the F1 population. Moreover, the lower resolution power of the silver-staining detection system may also reduce the number of available segregation markers (Wang et al. 2015). On the other hand, the number of LGs in this map was far more than the haploid chromosome number (n) of 11 (2n = 22) of Taxodium. This could be associated with an insufficient quantity of polymorphic markers linked on the map due to the absence of more intermediate loci, leading to gaps that divide chromosomes into several LGs (2 triples and 14 pairs; Guo et al. 2014; Lin et al. 2009). Reports concerning the number of LGs detected are greater than the number of plant chromosomes in several previous studies (Scotti et al. 2005; Komulainen et al. 2003; Shepherd et al. 2003; Iwata et al. 2001; Guo et al. 2014). Scotti et al. (2005) reported a genetic linkage map of P. abies (n = 12), which was comprised of 27 LGs containing at least four markers. In 2003, a GL map comprising 27 LGs of P. elliottii var. elliottii, and a map containing 23 LGs of P. caribaea var. hondurensis were constructed by Shepherd et al. (2003). These observations might be related to the type and number of individuals of the population, or the type and number of markers used in the mapping (Guo et al. 2014). In addition, LGs of maps typically do not correspond to the haploid chromosome numbers may be because of the nonrandom genomic distribution of different marker types and recombination rates between mapping parents on some chromosomes (Scotti et al. 2005; Cai et al. 2015).
QTL mapping
The identification of economically important QTLs is a significant foundation for MAS to improve trees and the studies of molecular regulations involved in the various characteristics. The present study represents the first QTL investigation of the growth traits of SH, BD and CW in Taxodium. QTLs associated with growth traits have been studied widely in many conifer species (Nowicka et al. 2013; Lepoittevin et al. 2012; Yoshimaru et al. 1998; Ukrainetz et al. 2008; Pelgas et al. 2011; Wheelerl et al. 2005). Ukrainetz et al. (2008) detected two and one QTLs for tree height and diameter at breast height, respectively, in Douglas-fir. Nowicka et al. (2013) successfully searched QTLs related to growth traits of diameter at breast height, of tree height, the number of needles per 10 cm shoots from the apical bud, needle width, needle length and needle area of P. sylvestris. Pelgas et al. (2011) identified 137 single QTLs related to growth and phenology, including 33 for bud flush, 52 for bud set and 52 for growth of Picea glauca.
Tang et al. (2015) showed that many QTL intervals controlling different fiber quality traits overlapped in some common chromosomal regions. In our study, four major QTLs on LG7 and LG13 were detected (Table 6; Figs. 1, 2, 3), of which one QTL was related significantly to SH (qtSH-1), one to BD (qtBD-1) and one to CW (qtCW-1), which were allocated to the same position at 2.695 cM on LG7, and explained 24.9, 27 and 21.7 % of the total variation of the three growth traits, respectively. The very high correlations among the traits (Table 5), the identical locations of three QTLs and the large phenotypic variances explained suggests that they may be early-growth traits mostly affected by seed size (or amount of storage tissue available to the developing embryo) (Escudero et al. 2000; Sexton et al. 1997). Moreover, these co-localized QTLs may be controlled by pleiotropic genes, which play important roles in the development of growth traits. However, whether the three traits are controlled by the same gene or different genes can still not be determined due to sketchy maps having insufficient markers. Therefore, more markers and a high-density detailed genetic linkage map is needed. The nearest marker of the three QTLs in LG7 was a distorted marker (Me14-Em8-170), and the map distance between this loci and those QTLs was only 1 cM. Previous studies reported that markers having segregation distortion were recognized as potentially powerful evolutionary forces (Cai et al. 2015) and may be associated with several QTLs (Chen et al. 2010a; Xu 2008; Luo et al. 2005).
Changes in biological and climatic factors across years may cause a bias in the phenotypic value assessment of quantitative traits (Chen et al. 2015). In this study, due to the QTLs being detected only in a single year, and with only one individual per genotype of the F1 population, there might be instability in this QTL in multiple years and a potential inconsistency in phenotypic assessments between the seedling stage and maturity. Despite addressing these problems, there have been complications in woody plants, and several measures, such as analyzing each of the traits for at least 3 consecutive years and increasing more than three replicates per genotype, are currently underway to ensure that QTLs detected were considered stable and allow us to employ markers in tree breeding accurately and effectively.
Conclusions
A genetic linkage map was constructed for T. ‘Zhongshansa’ using SRAP and EST-SSR markers. A total of 179 markers were distributed to 34 LGs with an observed map length of 976.5 cM and a mean distance of 7.0 cM between markers. In addition, four QTLs related to the growth traits of SH, BD and CW were detected based on the map constructed. Further, it is anticipated that a detailed analysis of QTL locations based on high-density saturated linkage maps of Taxodium will be a future task. Additionally, efforts to map more economically important traits, such as growth traits, wood quality and quantitative resistance, which segregate in the T. ‘Zhongshansa’ population, are also in progress.
References
Acheré V, Faivre-Rampant P, Jeandroz S, Besnard G, Markussen T, Aragones A, Fladung M, Ritter E, Favre J-M (2004) A full saturated linkage map of Picea abies including AFLP, SSR, ESTP, 5S rDNA and morphological markers. Theor Appl Genet 108:1602–1613
Alwala S, Kimbeng CA, Veremis JC, Gravois KA (2008) Linkage mapping and genome analysis in a Saccharum interspecific cross using AFLP, SRAP and TRAP markers. Euphytica 164:37–51
Cai CF, Cheng FY, Wu J, Zhong Y, Liu GX (2015) The first high-density genetic map construction in tree peony (Paeonia Sect. Moutan) using genotyping by specific-locus amplified fragment sequencing. PLoS ONE 10(5):e0128584
Canli FA (2004) Development of a second generation genetic linkage map for sour cherry using SSR markers. Pak J Biol Sci 7(10):1676–1683
Chakravarti A, Lasher LK, Reefer JE (1991) A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128:175–182
Chen MM, Feng FG, Sui X, Han SG (2010a) Genetic linkage maps of Pinus koraiensis Sieb. et Zucc. based on AFLP markers. Afr J Biotechnol 9(35):5659–5664
Chen MM, Feng FG, Sui X, Li MH, Zhao D, Han SJ (2010b) Construction of a framework map for Pinus koraiensis Sieb. et Zucc. using SRAP, SSR and ISSR markers. Trees 24:685–693
Chen J, Wang N, Fang LC, Liang ZC, Li SH, Wu BH (2015) Construction of a high-density genetic map and QTLs mapping for sugars and acids in grape berries. BMC Plant Biol 15:28
Cheng YL, Yang Y, Wang ZY, Qi BY, Yin YL (2015) Development and characterization of EST-SSR markers in Taxodium ‘zhongshansa’. Plant Mol Biol Rep 33:1804–1814
Creech D, Zhou LJ, Yin YL, Eguiluz-Piedra T (2011) Can Taxodium be improved? Arnoldia 69(2):11–20
Denny GC, Arnold MA (2007) Taxonomy and nomenclature of baldcypress, pondcypress, and montezuma cypress: one, two, or three species? Horttechnology 17(1):125–127
Eckert AJ, Pande B, Ersoz ES, Wright MH, Rashbrook VK, Nicolet CM, Neale DB (2009) High-throughput genotyping and mapping of single nucleotide polymorphisms in loblolly pine (Pinus taeda L.). Tree Genet Genomes 5:225–234
Escudero A, Nunez Y, Perez-Garcia F (2000) Is fire a selective force of seed size in pine species? Acta Oecol 21:245–256
García M, Villalba P, Acuña C, Oberschelp J, Harrand L, Surenciski M, Martínez M, Petroli C, Sansaloni C, Faria D, Grattapaglia D, Poltri SM (2011) A genetic linkage map for a Full sib population of Eucalyptus grandis using SSR, DArT, CG-SSR and EST-SSR markers. BMC Proceed 5(suppl 7):P26
Gaudet M, Jorge V, Paolucci I, Beritognolo I, Scarascia Mugnozza G, Sabatti M (2008) Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet Genomes 4:25–36
Guo HL, Ding WW, Chen JB, Chen X, Zheng YQ, Wang ZY, Liu JX (2014) Genetic linkage map construction and QTL mapping of salt tolerance traits in Zoysiagrass (Zoysia japonica). PLoS ONE 9(9):e107249
He ZX, Shi JS, Wang MX, Yu RZ, Chen XC (2000) The construction of molecular linkage map in Chinese fir. J Nanjing For Univ 24(6):22–26
Hu JB, Zhou XY, Li JW (2010) Development of novel EST-SSR markers for cucumber (Cucumis sativus) and their transferability to related species. Sci Hortic 125:534–538
Huang H, Lu J, Ren Z, Hunter W, Dowd SE, Dang P (2011) Mining and validating grape (Vitis L.) ESTs to develop EST-SSR markers for genotyping and mapping. Mol Breed 28:241–254
Iwata H, Ujino-Ihara T, Yoshimura K, Nagasaka K, Mukai Y, Tsumura Y (2001) Cleaved amplified polymorphic sequence markers in sugi, Cryptomeria japonica D. Don, and their locations on a linkage map. Theor Appl Genet 103:881–895
Jena KK, Jeung JU, Lee JH, Choi HC, Brar DS (2006) High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t), and marker-assisted selection for BPH resistance in rice (Oryza sativa L.). Theor Appl Genet 112:288–297
Jermstad KD, Eckert AJ, Wegrzyn JL, Delfino-Mix A, Davis DA, Burton DC, Neale DB (2011) Comparative mapping in Pinus: sugar pine (Pinus lambertiana Dougl.) and loblolly pine (Pinus taeda L.). Tree Genet Genomes 7:457–468
Kang BY, Mann IK, Major JE, Rajora OP (2010) Near-saturated and complete genetic linkage map of black spruce (Picea mariana). BMC Genom 11:515
Kiss GB, Csanádi G, Kálmán K, Kaló P, Ǒkrész L (1993) Construction of a basic genetic map for alfalfa using RFLP, RAPD, isozyme and morphological markers. Mol Gen Genet 238:129–137
Komulainen P, Brown GR, Mikkonen M, Karhu A, García-Gil MR, Malley DO, Lee B, Neale DB, Savolainen O (2003) Comparing EST-based genetic maps between Pinus sylvestris and Pinus taeda. Theor Appl Genet 107:667–678
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lepoittevin C, Harvengt L, Plomion C, Garnier-Géré P (2012) Association mapping for growth, straightness and wood chemistry traits in the Pinus pinaster Aquitaine breeding population. Tree Geneti Genomes 8:113–126
Lerceteau E, Plomion C, Andersson B (2000) AFLP mapping and detection of quantitative trait loci (QTLs) for economically important traits in Pinus sylvestris: a preliminary study. Mol Breed 6:451–458
Lerceteau E, Szmidt AE, Andersson B (2001) Detection of quantitative trait loci in Pinus sylvestris L. across years. Euphytica 121:117–122
Leroy T, De Bellis F, Legnate H, Kananura E, Gonzales G, Pereira LF, Andrade AC, Charmetant P, Montagnon C, Cubry P, Marrccini P, Pot D, de Kochko A (2011) Improving the quality of African robustas: QTLs for yield- and quality-related traits in Coffea canephora. Tree Genet Genomes 7:781–798
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Lin ZX, Zhang YX, Zhang XL, Guo XP (2009) A high-density integrative linkage map for Gossypium hirsutum. Euphytica 166:35–45
Liu YC, Liu S, Liu DC, Wei YX, Liu C, Yang YM, Tao CG, Liu WS (2014) Exploiting EST databases for the development and characterization of EST-SSR markers in blueberry (Vaccinium) and their cross-species transferability in Vaccinium spp. Sci Hortic 176:319–329
Luo L, Zhang YM, Xu S (2005) A quantitative genetics model for viability selection. Heredity 94:347–355
Marone D, Laido G, Gadaleta A, Colasuonno P, Ficco DBM, Giancaspro A, Giove S, Panio Giosué, Russo MA, Vita PD, Cattivelli L, Papa R, Blanco A, Mastrangelo AM (2012) A high-density consensus map of A and B wheat genomes. Theor Appl Genet 125:1619–1638
Martínez-García PJ, Stevens KA, Wegrzyn JL, Liechty J, Crepeau M, Langley CH, Neale DB (2013) Combination of multipoint maximum likelihood (MML) and regression mapping algorithms to construct a high-density genetic linkage map for loblolly pine (Pinus taeda L.). Tree Genet Genomes 9:1529–1535
Moraga-Suazo P, Orellana L, Quiroga P, Balocchi C, Sanfuentes E, Whetten RW, Hasbún R, Valenzuela S (2014) Development of a genetic linkage map for Pinus radiata and detection of pitch canker disease resistance associated QTLs. Trees 28:1823–1835
Moriguchi Y, Ujino-Ihara T, Uchiyama K, Futamura N, Saito M, Ueno S, Matsumoto A, Tani N, Taira H, Shinohara K, Tsumura Y (2012) The construction of a high-density linkage map for identifying SNP markers that are tightly linked to a nuclear-recessive major gene for male sterility in Cryptomeria japonica D. Don. BMC Genomics 13:95
Muchero W, Guo JJ, DiFazio SP, Chen JG, Ranjan P, Slavov GT, Gunter LE, Jawdy S, Bryan AC, Sykes R, Ziebell A, Kápstě J, Porth L, Skyba O, Unda F, El-Kassaby YA, Douglas CJ, Mansfield SD, Martin J, Schackwitz W, Evans LM, Czarnecki O, Tuskan GA (2015) High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC Genom 16:24
Mukai Y, Suyama Y, Tsumura Y, Kawahara T, Yoshimaru H, Kondo T, Tomaru N, Kuramoto N, Murai M (1995) A linkage map for sugi (Cryptomeria japonica) based on RFLP, RAPD, and isozyme loci. Theor Appl Genet 90:835–840
Nikaido AM, Ujino T, Iwata H, Yoshimura K, Yoshimura H, Suyama Y, Murai M, Nagasaka K, Tsumura Y (2000) AFLP and CAPS linkage maps of Cryptomeria japonica. Theor Appl Genet 100:825–831
Nodari RO, Tsai SM, Gilbertson RL, Gepts P (1993) Towards an integrated linkage map of common bean 2. Development of an RFLP-based linkage map. Theor Appl Genet 85:513–520
Nowicka A, Ukalska J, Simińska J, Szyp-Borowska I (2013) Characterization and mapping of QTL used in breeding of Scots pine (Pinus sylvestris L.). Folia Forestalia Polonica 55(4):168–173
Pelgas B, Bousquet J, Beauseigle S, Isabe N (2005) A composite linkage map from two crosses for the species complex Picea mariana × Picea rubens and analysis of synteny with other Pinaceae. Theor Appl Genet 111:1466–1488
Pelgas B, Bousquet J, Meirmans PG, Ritland K, Isabel N (2011) QTL mapping in white spruce: gene maps and genomic regions underlying adaptive traits across pedigrees, years and environments. BMC Genom 12:145
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P (2006) SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol Gen Genomics 276:436–449
Pot D, Rodrigues JC, Rozenberg P, Chantre G, Tibbits J, Cahalan C, Pichavant F, Plomion C (2006) QTLs and candidate genes for wood properties in maritime pine (Pinus pinaster Ait.). Tree Genet Genomes 2:10–24
Qi BY, Yang Y, Yin YL, Xu M, Li HG (2014) Denovo sequencing, assembly, and analysis of the Taxodium ‘Zhongshansa’ roots and shoots transcriptome in response to short-term waterlogging. BMC Plant Biol 14:201
Qin HD, Guo WZ, Zhang YM, Zhang TZ (2008) QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor Appl Genet 117:883–894
Rieseberg LH, Baird S, Gardner K (2000) Hybridisation, introgression, and linkage evolution. Plant Mol Biol 42:205–224
Rittera E, Aragonésa A, Markussenb T, Acheréc V, Espinela S, Fladung M, Wrobel S, Faivre-Rampant P, Jeandroz S, Favre JM (2002) Towards construction of an ultra high density linkage map for Pinus pinaster. Ann For Sci 59:637–643
Scotti I, Burelli A, Cattonaro F, Chagné D, Fuller J, Hedley PE, Jansson G, Lalanne C, Madur D, Neale D, Plomion C, Powell W, Troggio M, Morgante M (2005) Analysis of the distribution of marker classes in a genetic linkage map: a case study in Norway spruce (Picea abies karst). Tree Genet Genomes 1:93–102
Sewell MM, Davis MF, Tuskan GA, Wheeler NC, Elam CC, Bassoni DL, Neale DB (2002) Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). II. Chemical wood properties. Theor Appl Genet 104:214–222
Sexton PJ, Peterson CM, Boote KJ, White JW (1997) Early-season growth in relation to region of domestication, seed size, and leaf traits in common bean. Field Crops Res 52:69–78
Shepherd M, Cross M, Dieters MJ, Henry R (2003) Genetic maps for Pinus elliottii var. elliottii and P. caribaea var. hondurensis using AFLP and microsatellite markers. Theor Appl Genet 106:1409–1419
Sun SJ, Gao W, Lin SQ, Zhu J, Xie BG, Lin ZB (2006) Analysis of genetic diversity in Ganoderma population with a novel molecular marker SRAP. Appl Microbiol Biotechnol 72:537–543
Tang SY, Teng ZH, Zhai TF, Fang XM, Liu F, Liu DJ, Zhang J, Liu DX, Wang SF, Zhang K, Shao QS, Tan ZY, Paterson AH, Zhang ZS (2015) Construction of genetic map and QTL analysis of fiber quality traits for Upland cotton (Gossypium hirsutum L.). Euphytica 201:195–213
Temesgen B, Brown GR, Harry DE, Kinlaw CS, Sewell MM, Neale DB (2001) Genetic mapping of expressed sequence tag polymorphism (ESTP) markers in loblolly pine (Pinus taeda L.). Theor Appl Genet 102:664–675
Tong CF, Shi JS (2004) Construction genetic linkage maps in chinese fir using F1 progeny. Acta Genetica Sinica 31(10):1149–1156
Tsumura Y, Yoshimura K, Tomaru N, Ohba K (1995) Molecular phylogeny of conifers using RFLP analysis of PCR-amplified specific chloroplast genes. Theor Appl Genet 91:1222–1236
Ukrainetz NK, Ritland K, Mansfield SD (2008) Identification of quantitative trait loci for wood quality and growth across eight full-sib coastal Douglas-fir families. Tree Genet Genomes 4:159–170
Van Ooijen JW (2004) MapQTL 5.0: software for the mapping quantitative trait loci in experimental populations. Plant Research International, Wageningen
Van Ooijen JW (2006) JoinMap 4.0, software for the calculation of genetic linkage maps. Kyazama BV, Wageningen
Voorrips RE (2002) Mapchart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Wang G, Pan J, Li XZ, He H, Wu AZ, Run C (2005) Construction of a cucumber genetic linkage map with SRAP markers and location of the genes for lateral branch traits. Sci China Life Sci 48(3):213–220
Wang ZY, Cheng YL, Yin YL, Yu CG, Li HG (2015) The cross-family transferability of SSR markers developed from the transcriptome of Taxodium ‘Zhongshanshan 406’ and its application in genotypic assessment. Mol Plant Breed 13(7):1631–1638
Wheelerl NC, Jermstad KD, Krutovsky K, Aitken SN, Howe GT, Krakowski J, Neale DB (2005) Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. IV. Cold-hardiness QTL verification and candidate gene mapping. Mol Breed 15:145–156
Xu SZ (2008) Quantitative trait locus mapping can benefit from segregation distortion. Genetics 180:2201–2208
Yang HX, Luo R, Zhao FC, Liu TY, Liu CX, Huang SW (2013a) Constructing genetic linkage maps for Pinus elliottii var. elliottii and Pinus caribaea var. hondurensis using SRAP, SSR, EST and ISSR markers. Trees 27:1429–1442
Yang J, Dai PF, Zhou TH, Huang ZH, Feng L, Su HL, Liu ZL, Zhao GF (2013b) Genetic diversity and structure of wintersweet (Chimonanthus praecox) revealed by EST-SSR markers. Sci Hortic 150:1–10
Yao JX, Li HG, Ye J, Shi LL (2016) Relationship between parental genetic distance and offspring’s heterosis for early growth traits in Liriodendron: implication for parent pair selection in cross breeding. New Forest 47:163–177
Yin TM, Wang XR, Andersson B, Lerceteau-Kǒhler E (2003) Nearly complete genetic maps of Pinus sylvestris L. (Scots pine) constructed by AFLP marker analysis in a full-sib family. Theor Appl Genet 106:1075–1083
Yoshimaru H, Ohba K, Tsurumi K, Tomaru N, Murai M, Mukai Y, Suyama Y, Tsumura Y, Kawahara T, Sakamaki Y (1998) Detection of quantitative trait loci for juvenile growth, flower bearing and rooting ability based on a linkage map of sugi (Cryptomeria japonica D. Don). Theor Appl Genet 97:45–50
Yu CG, Yin YL, Xu JH (2009) Identification of Taxodium hybrids by SRAP analysis. Sci Silvae Sinicae 45(2):142–146
Zhang ZS, Hu MC, Zhang J, Liu DJ, Zheng J, Zhang K, Wang W, Wan Q (2009) Construction of a comprehensive PCR-based marker linkage map and QTL mapping for fiber quality traits in upland cotton (Gossypium hirsutum L.). Mol Breed 24:49–61
Zhong S, Toubia-Rahme H, Steffenson BJ, Smith KP (2006) Molecular mapping and marker-assisted selection of genes for septoria speckled leaf blotch resistance in barley. Phytopathology 96(9):993–999
Zhou LJ, Creech DL, Krauss KW, Yin YL, Kulhavy DL (2010) Can we improve the salinity tolerance of genotypes of Taxodium by using varietal and hybrid crosses? HortScience 45(12):1773–1778
Zhou DQ, Mo HB, Lu ZG, Yu CG, Xu HB, Yin YL (2012) Genetic diversity analysis of superior individuals of Taxodium mucronatum based on SRAP marker. J Plant Resourc Environ 21(1):36–41
Authors’ contributions
ZYW and YLC performed the experiments, analyzed the data and wrote the manuscript. CGY, YY, QS and ZYH participated in DNA extraction, genotyping and statistical analysis. HGL and YLY conceived the idea, proposed, initiated and led the project, interpreted scientific information and participated in manuscript preparation. All authors read and approved the final manuscript.
Acknowledgements
This study was financially supported by grants from the National Natural Science Foundation of China (31570593), the Natural Science Foundation of Jiangsu Province (BK20150551) and the Program of Innovation Capacity Construction of Jiangsu Province (BM2015019).
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, Z., Cheng, Y., Yin, Y. et al. Genetic linkage map construction and QTL mapping of seedling height, basal diameter and crown width of Taxodium ‘Zhongshanshan 302’ × T. mucronatum . SpringerPlus 5, 936 (2016). https://doi.org/10.1186/s40064-016-2617-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-2617-3