Abstract
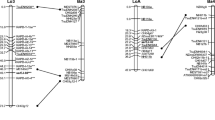
A total of 122 F1 individuals from a single full-sib interspecific hybrid family crossed between Pinus elliottii var. elliottii (PEE) and P. caribaea var. hondurensis (PCH) were used to construct a detailed genetic linkage maps using four types of molecular markers: sequence-related amplified polymorphism (SRAP), microsatellite (SSR), expressed sequence tag polymorphism (ESTP) and inter-simple sequence repeat (ISSR). There were 381 SRAP, 108 SSR, 25 ESTP and 32 ISSR loci, segregating in the interspecific F1 hybrid individuals. Framework maps were constructed at a LOD score threshold of 4.0 using the JoinMap® 3.0. The map for the male parent (PCH) had 108 markers in 16 linkage groups (LGs), with a total length of 1,065.9 cM (Kosambi) and an average marker interval of 9.87 cM. The map for the female parent (PEE) contained 93 markers in 19 LGs, with a total length of 1,006.7 cM (Kosambi) and an average marker interval of 10.82 cM. The maps for PCH and PEE covered 56.5 and 70.3 % of their respective genomes. Based on the position of 36 loci segregating in both parents, 8 homologous LGs between PEE and PCH were identified.






Similar content being viewed by others
References
Ahmad R, Potter D, Southwick SM (2004) Genotyping of peach and nectarine cultivars with SSR and SRAP molecular markers. J Am Soc Hortic Sci 129:204–210
Alwala S, Kimbeng CA, Veremis JC et al (2008) Linkage mapping and genome analysis in a Saccharum interspecific cross using AFLP, SRAP and TRAP markers. Euphytica 164:37–51
Andersson F (2005) Ecosystems of the world: coniferous forests. Elsevier, Amsterdam
Auckland L, Bui T, Zhou Y et al (2002) Conifer microsatellite handbook. Texas A&M, College Station
Brown GR, Kadel EE, Bassoni DL et al (2001) Anchored reference loci in loblolly pine (Pinus taeda L.) for integrating pine genomics. Genetics 159:799–809
Chancerel E, Lepoittevin C, Le Provost G et al (2011) Development and implementation of a highly-multiplexed SNP array for genetic mapping in maritime pine and comparative mapping with loblolly pine. BMC genomics 12(1):368. doi:10.1186/1471-2164-12-368.
Chen MM, Feng FJ, Sui X et al (2010) Construction of a framework map for Pinus koraiensis Sieb. et Zucc. using SRAP, SSR and ISSR markers. Trees 24:685–693
Couch JA, Fritz PJ (1990) Isolation of DNA from plant high in polyphenolic. Plant Mol Biol Rep 8:8–12
Dale G (1994) Genetic mapping in an interspecific hybrid between Pinus caribaea and Pinus elliottii, Unpublished PhD thesis, University of Queensland, Brisbane
Devey ME, Bell JC, Smith DN et al (1996) A genetic linkage map for Pinus radiata based on RFLP, RAPD and microsatellite markers. Theor Appl Genet 92:673–679
Devey ME, Sewell MM, Uren TL et al (1999) Comparative mapping in loblolly and radiata pine using RFLP and microsatellite markers. Theor Appl Genet 99:656–662
Devey ME, Bell JC, Moran G (2001) A set of microsatellite markers for fingerprinting and breeding applications in Pinus radiata. Genome 45:984–989
Echt CS, Deverno LL, Anzidei M et al (1998) Chloroplast microsatellite reveal population genetic diversity in red pine, Pinus resinosa Ait. Mol Ecol 7:307–316
Echt CS, Saha S, Krutovsky KV et al (2011) An annotated genetic map of loblolly pine based on microsatellite and cDNA markers. BMC genetics 12(1): 17. doi:10.1186/1471-2156-12-17.
Eckert AJ, van Heerwaarden J, Wegrzyn JL et al (2010) Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae). Genetics 185(3):969–982
Esposito MA, Martin EA, Cravero VP et al (2007) Characterization of pea accessions by SRAP’s markers. Sci Hortic 113:329–335
Ferriol M, Picó B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282
Ferriol M, Picó B, Nuez F (2004) Morphological and molecular diversity of a collection of Cucurbita maxima landraces. J Am Soc Hortic Sci 129:60–69
Guo DL, Luo ZR (2006) Genetic relationships of some PCNA persimmons (Diospyros kaki Thunb.) from China and Japan revealed by SRAP analysis. Genet Res Crop Evol 53:1603–1797
Haines RJ (2000) Clonal forestry in Queensland and implications for hybrid breeding strategies. In: Dungey HS, Dieters MJ, Nikles DG (compilers) Hybrid breeding and genetics of forest trees. Proc QFRI/CRC-SPF Symp. Department of Primary Industries, Brisbane, pp 386–389
Harry DE, Temesgen B, Neale DB (1998) Codominant PCR-based markers for Pinus taeda developed from mapped cDNA clones. Theor Appl Genet 97:327–336
Huang YQ, Zhao FC (2007) Current situation and developing proposal on the breedling of Pinus elliottii × P. caribaea hybrid in Guangdong (in Chinese). J Fujian For Sci Tech 34(1):158–163
Jermstad KD, Eckert AJ, Wegrzyn JL et al (2010) Comparative mapping in Pinus: sugar pine (Pinus lambertiana Dougl.) and loblolly pine (Pinus taeda L.). Tree Genet Genomes 7:457–468
Kinlaw CS, Neale DB (1997) Complex gene families in pine genomes. Trends Plant Sci 2:356–359
Komulainen P, Brown GR, Mikkonen M et al (2003) Comparing EST-based genetic maps between Pinus sylvestris and Pinus taeda. Theor Appl Genet 107:667–678
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Krutovsky KV, Troggio M, Brown GR et al (2004) Comparative mapping in the Pinaceae. Genetics 168(1):447–461
Kuang H, Richardson T, Carson S, Wilcox P, Bongarten B (1999) Genetic analysis of inbreeding depression in plus tree 850.55 of Pinus radiata D. Don. I. Genetic map with distorted markers. Theor Appl Genet 98:697–703
Kubisiak TL, Nelson CD, Nance WL et al (1995) RAPD linkage mapping in a Longleaf pine × slash pine F1 family. Theor Appl Genet 90:1119–1127
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Lin ZX, He DH, Zhang XL et al (2005) Linkage map construction and mapping QTL for cotton fibre quality using SRAP, SSR and RAPD. Plant Breed 124:180–187
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev. Genetics 12(2):111–122. Nature Publishing Group. doi:10.1038/nrg2931
Nelson CD, Nance WL, Doudrick RL (1993) A partial genetic linkage map of slash pine (Pinus elliottii Engelm. var. elliottii) based on random amplified polymorphic DNAs. Theor Appl Genet 87:145–151
Nelson CD, Kubisiak TL, Stine M et al (1994) A genetic linkage map of Longleaf pine (Pinus palustris Mill.) based on random amplified polymorphic DNAs. J Hered 85(6):433–439
Nikes DG (1979) Genetic improvement of low land tropical conifers. FAO, Rome
Nikles DG (1995) Hybrid of the slash-Caribbean-Central American pine complex: characteristics, bases of superiority and potential utility in South China and elsewhere. In: Shen X (ed) Proceedings: forest tree improvement in the Asia-Pacific region. China Forestry Publishing House, Beijing, pp 168–186
Perry DJ, Furnier GR (1996) Pinus banksiana has at least seven expressed alcohol dehydrogenase genes in two linked groups. Proc Natl Acad Sci USA 93:13020–13023
Remington DL, Whetten RW, Liu BH et al (1999) Construction of an AFLP genetic map with nearly complete genome coverage in Pinus taeda. Theor Appl Genet 98:1279–1292
Riaz S, Dangl GS, Edwards KJ et al (2004) A microsatellite marker based framework linkage map of Vitis vinifera L. Theor Appl Genet 108:864–872
Rieseberg LH, Baird S, Gardner K (2000) Hybridisation, introgression, and linkage evolution. Plant Mol Biol 42:205–224
Ritland K, Krutovsky K, Tsumura Y et al (2011) Genetic mapping in conifers. In: Genetics, genomics and breeding of conifers. In: Plomion C, Bousquet J, Enfield KC (eds) CRC Press Science Publishers, Boca Raton, pp 196–238
Ruiz JJ, García-Martínez S, Picó B et al (2005) Genetic variability and relationship of closely related Spanish traditional cultivars of tomato as detected by SRAP and SSR markers. J Am Soc Hortic Sci 130:88–94
Shepherd M, Cross M, Dieters MJ et al (2003) Genetic maps for Pinus elliottii var. elliottii and P. caribaea var. hondurens is using AFLP and microsatellite markers. Theor Appl Genet 106:1409–1419
Temesgen T, Brown GR, Harry DE et al (2001) Genetic mapping of expressed sequence tag polymorphism (ESTp) markers in loblolly pine (pinus taeda L). Theor Appl Genet 102:664–675
Uzun A, Yesiloglu T et al (2009) Genetic diversity and relationships within Citrus and related genera based on sequence related amplified polymorphism markers (SRAPs). Sci Hortic 121:306–312
Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageninggen, The Netherlands
Voorrips RE (2002) Mapchart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang H, Malcolm DC, Fletcher AM (1999) Pinus caribaea in China: introduction, genetic resources and future prospects. For Ecol Manage 171:1–15
White TL, Powell G, Rockwood D, Parker S (eds) (1996) Cooperative forest genetics research program 38th Annual progress report. School of Forest Resources and Conservation, IFAS, Univ. Florida, Gainesville, FL
Zhang ZS, Hu MC (2009) Construction of a comprehensive PCR-based marker linkage map and QTL mapping for fiber quality traits in upland cotton (Gossypium hirsutum L.). Mol Breed 24:49–61
Zheng YQ, Wang HR (2000) Genetic resources and breeding of Pinus caribaea in China. In: Forests and society: the role of research, abstracts of group discussions. XX IUFRO World Congress
Zhou Y, Gwaze DP, Bui T et al (2003) No clustering for linkage map based on low-copy and undermethylated microsatellites. Genome 45:809–816
Acknowledgments
The authors thank J. Lin and X. Lai of the Longshan Forest Farm, Guangdong Academy of Forestry for assistance in collection of the samples. Drs. Harry Wu and Milos Ivkovich of CSIRO Plant Industry, Australia are thanked for valuable comments and English editing on an early draft.
The study reported here was financially supported by the National Natural Science Foundation of China (30671706).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. E. Carlson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Hx., Luo, R., Zhao, Fc. et al. Constructing genetic linkage maps for Pinus elliottii var. elliottii and Pinus caribaea var. hondurensis using SRAP, SSR, EST and ISSR markers. Trees 27, 1429–1442 (2013). https://doi.org/10.1007/s00468-013-0890-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-013-0890-0