Abstract
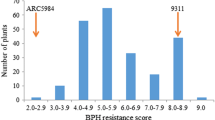
The brown planthopper (BPH), Nilaparvata lugens Stål, is one of the most destructive pests to the rice production in the world. Thus, there is an urgency to identify new resistant genes for breeding. AC-1613 is an indica variety that has been reported to confer broad-spectrum resistance to BPH. In the present study, we found that AC-1613 exhibited strong antibiosis towards BPH insects. The body weight was significantly decreased when the insects fed on AC-1613 plants. By using BPH weight gain as an index of phenotyping, a novel dominant locus for resistance to BPH, designed as Bph30, was identified and its near-isogenic line (NIL) in 9311 background was developed. The F2 population derived from a cross between AC-1613 and 9311 was used for mapping the gene. Through QTL scan, we located the gene on the short arm of chromosome 4 between RM16278 and RM16425, which explained 42.7% of the phenotypic variance (PEV) of BPH resistance in the F2 population. The gene was finally located in a region flanking by simple sequence repeat (SSR) markers SSR-28 and SSR-69 through high-resolution mapping, the distance between the two markers in Nipponbare genome is 37.5 kb. In addition, SSR markers RM16294 and RM16299 tightly linked to Bph30 were applied effectively in introgressing Bph30 into elite rice cultivars. The developed NILs showed a strong antibiosis and high resistance to BPH.





Similar content being viewed by others
References
Alam SN, Cohen MB (1998) Detection and analysis of QTLs for resistance to the brown planthopper, Nilaparvata lugens, in a doubled-haploid rice population. Theor Appl Genet 97:1370–1379
Backus EA, Serrano MS, Ranger CM (2005) Mechanisms of hopperburn: an overview of insect taxonomy, behavior, and physiology. Annu Rev Entomol 50:125–151
Chen JW, Wang L, Pang XF, Pan QH (2006) Genetic analysis and fine mapping of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene bph19(t). Mol Gen Genomics 275:321–329
Du B, Zhang WL, Liu BF, Hu J, Wei Z, Shi ZY, He RF, Zhu LL, Chen RZ, Han B, He GC (2009) Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci U S A 106:22163–22168
Guo JP, Xu CX, Wu D, Zhao Y, Qiu YF, Wang XX, Ouyang YD, Cai BD, Liu X, Jing SL, Shangguan XX, Wang HY, Ma YH, Hu L, Wu Y, Shi SJ, Wang WL, Zhu LL, Xu X, Chen RZ, Feng YQ, Du B, He GC (2018) Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat Genet 50:297–306
Hao PY, Liu CX, Wang YC, Chen RZ, Tang M, Bo D, Zhu LL, He GC (2008) Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol 146:1810–1820
He J, Liu YQ, Liu YL, Jiang L, Wu H, Kang HY, Liu SJ, Chen LM, Liu X, Cheng XN, Wan JM (2012) High-resolution mapping of brown planthopper (BPH) resistance gene Bph27(t) in rice (Oryza sativa L). Mol Breed 31:549–557
Hu J, Xiao C, Cheng MX, Gao GJ, Zhang QL, He YQ (2015a) Fine mapping and pyramiding of brown planthopper resistance genes QBph3 and QBph4 in an introgression line from wild rice O. officinalis. Mol Breed 35:3
Hu J, Xiao C, Cheng MX, Gao GJ, Zhang QL, He YQ (2015b) A new finely mapped Oryza australiensis-derived QTL in rice confers resistance to brown planthopper. Gene 561:132–137
Huang Z, He GC, Shu LH, Li XH, Zhang QF (2001) Identification and mapping of two brown planthopper resistance genes in rice. Theor Appl Genet 102:929–934
Ishii T, Brar DS, Multani DS, Khush GS (1994) Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa. Genome 37:217–221
Jairin JP, Phengrat K, Teangdeerith SG, Vanavichit A, Toojinda T (2006) Mapping of a broad-spectrum brown planthopper resistance gene, Bph3, on rice chromosome 6. Mol Breed 19:35–44
Jairin J, Sansen K, Wongboon W, Kothcharerk J (2010) Detection of a brown planthopper resistance gene bph4 at the same chromosomal position of Bph3 using two different genetic backgrounds of rice. Breeding Sci 60:71–75
Jena KK, Jeung JU, Lee JH, Choi HC, Brar DS (2006) High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t), and marker-assisted selection for BPH resistance in rice (Oryza sativa L). Theor Appl Genet 112:288–297
Jing SL, Zhao Y, Bo D, Chen RZ, Zhu LL, He GC (2017) Genomics of interaction between the brown planthopper and rice. Curr Opin Insect Sci 19:82–87
Kharat KR, Sawant MV, Peter S, Hardikar BP (2010) Development and characterization of new cell line BPH22 from midgut epithelial cells of Poekilocerus pictus (Fabricius, 1775). In Vitro Cell Dev Biol-An 46:824–827
Kumar K, Sarao PS, Bhatia D, Neelam K, Kaur A, Mangat GS, Brar DS, Singh K (2018) High-resolution genetic mapping of a novel brown planthopper resistance locus, Bph34 in Oryza sativa L. X Oryza nivara (Sharma & Shastry) derived interspecific F2 population. Theor Appl Genet 131:1163–1171
Ling KC, Tiongco ER, Aguiero VM (1978) Rice ragged stunt, a new virus disease. Plant Disease Reporter 62:701–705
Liu GQ, Yan HH, Qiang F, Qian Q, Zhang ZT, Zhai WX, Zhu LH (2001) Mapping of a new gene for brown planthopper resistance in cultivated rice introgressed from Oryza eichingeri. Chinese Sci Bull 46:1459–1462
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A 21:9828–9832
Mohanty SK, Panda RS, Mohapatra SL, Nanda A, Behera L, Jena M, Sahu RK, Sahu SC, Mohapatra T (2017) Identification of novel quantitative trait loci associated with brown planthopper resistance in the rice landrace Salkathi. Euphytica 213:38
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Myint KKM, Fujita D, Matsumura M, Sonoda T, Yoshimura A, Yasui H (2012) Mapping and pyramiding of two major genes for resistance to the brown planthopper (Nilaparvata lugens [Stål]) in the rice cultivar ADR52. Theor Appl Genet 124:495–504
Pathak MD, Cheng CH, Fortuno ME (1969) Resistance to Nephotettix impicticeps and Nilaparvata lugens in varieties of rice. Nature 223:502–504
Pathak MD, Khush GS (1979) Studies of varietal resistance in rice to the brown planthopper at the international rice research institute. In: IRRI (ed) Brown planthopper: threat to rice production in Asia. IRRI, Los Banõs, Philippines, pp 285–301
Qiu YF, Guo JP, Jing SL, Zhu LL, He GC (2010) High-resolution mapping of the brown planthopper resistance gene Bph6 in rice and characterizing its resistance in the 9311 and Nipponbare near-isogenic backgrounds. Theor Appl Genet 121:1601–1611
Qiu YF, Guo JP, Jing SL, Zhu LL, He GC (2012) Development and characterization of japonica rice lines carrying the brown planthopper-resistance genes BPH12 and BPH6. Theor Appl Genet 124:485–494
Rahman ML, Jiang WZ, Chu SH, Qiao YL, Ham TH, Woo MO, Lee J, Khanam MS, Chin JH, Jeung JU, Brar DS, Jena KK, Koh HJ (2009) High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor Appl Genet 119:1237–1246
Ren JS, Gao FY, Wu XT, Lu XJ, Zeng LH, Lv JQ, Su XW, Luo H, Ren GJ (2016) Bph32, a novel gene encoding an unknown SCR domain-containing protein, confers resistance against the brown planthopper in rice. Sci Rep 6:37645
Rivera CT, Ou SH, Iida TT (1966) Grassy stunt disease of rice and its transmission by the planthopper Nilaparvata lugens Stal. Plant Disease Reporter 50:453–456
Sun LH, Su CC, Wang CM, Zhai HQ, Wan JM (2005) Mapping of a major resistance gene to the brown planthopper in the rice cultivar Rathu Heenati. Breeding Sci 55:391–396
Sun LH, Wang CM, Su CC, Liu YQ, Zhai HQ, Wan JM (2006) Mapping and marker-assisted selection of a brown planthopper resistance gene bph2 in rice (Oryza sativa L.). Acta Genet Sin 33:717–723
Van Ooijen JW (2006) JoinMap 4.0 Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V, Wageningen, Netherlands
Van Ooijen JW (2009) MapQTL 6.0 Software for the mapping of quantitative trait loci in experimental populations. Kyazma B.V, Wageningen, Netherlands. https://www.kyazma.nl/docs/MQ6Manual.pdf
Wang Y, Cao LM, Zhang YX, Cao CX, Liu F, Huang FK, Qiu YF, Li RB, Lou XJ (2015) Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice. J Exp Bot 66:6035–6045
Wang YC, Tang M, Hao PY, Yang ZF, Zhu LL, He GC (2008) Penetration into rice tissues by brown planthopper and fine structure of the salivary sheaths. Entomol Exp Appl 129:295–307
Watanabe T, Kitagawa H (2000) Photosynthesis and translocation of assimilates in rice plants following phloem feeding by the planthopper Nilaparvata lugens (homoptera: Delphacidae). J Econ Entomol 93:1192–1198
Yang HY, Ren X, Weng QM, Zhu LL, He GC (2002) Molecular mapping and genetic analysis of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene. Hereditas 136:39–43
Yang HY, You AQ, Yang ZF, Zhang FT, He RF, Zhu LL, He GC (2004) High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.). Theor Appl Genet 110:182–191
Yue B, Xue WY, Luo LJ, Xing YZ (2006) QTL analysis for flag leaf characteristics and their relationships with yield and yield traits in rice. Acta Genet Sin 33:824–832
Zhao Y, Huang J, Wang ZZ, Jing SL, Wang Y, Ouyang YD, Cai BD, Xin XF, Liu X, Zhang CX, PanYF MR, Li QF, Jiang WH, Zeng Y, Shangguan XX, Wang HY, Du B, Zhu LL, Xu X, Feng YQ, He SY, Chen RZ, Zhang QF, He GC (2016) Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc Natl Acad Sci U S A 113:12850–12855
Acknowledgements
This study was supported by grants from the National Program on Research & Development of Transgenic Plants (2016ZX08009-003-001), the National Natural Science Foundation of China (31630063), and the National Key Research and Development Program (2016YFD0100600).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
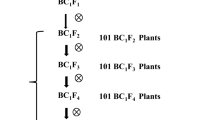
Fig. S1
The pedigree for developing mapping populations and NILs (PNG 70.1 kb)
Fig. S2
Overview of Parafilm sachet test of the BPH. a The distant view and b the close shot of Parafilm sachet test of the BPH. The labels with ‘L86’ and ‘L102’ denote recombinant individuals from mapping population (PNG 2483 kb)
Fig. S3
High-resolution mapping of Bph30 gene of part recombinants in BC2F2 populations. The upper local linkage map was constructed by seven SSR markers, scale bar = 20 kb. The below graphical presentation indicates the genotypes and phenotypes of the recombinants. The white and gray bars indicate the marker genotypes of 9311 homozygotes and heterozygotes, respectively. Weight gains indicate mean ± SD, n ≥ 3 repeats (PNG 123 kb)
Fig. S4
Genetic background assay of BC6F1 using the RICE6K array. The red lines indicate the SNP loci with homozygous AC-1613 genotypes and the blue lines indicate the SNP loci with heterozygous genotypes. The asterisk (*) denotes the position of Bph30 locus. (PNG 34.8 kb)
Table S1
Populations designated for identification and mapping of Bph30 (DOCX 18.9 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Shi, S., Guo, Q. et al. High-resolution mapping of a gene conferring strong antibiosis to brown planthopper and developing resistant near-isogenic lines in 9311 background. Mol Breeding 38, 107 (2018). https://doi.org/10.1007/s11032-018-0859-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0859-1