Abstract
Seed is one of the key factors of crop productivity. Therefore, a comprehension of the mechanisms underlying seed formation in cultivated plants is crucial for the quantitative and qualitative progress of agricultural production. In angiosperms, two pathways of reproduction through seed exist: sexual or amphimictic, and asexual or apomictic; the former is largely exploited by seed companies for breeding new varieties, whereas the latter is receiving continuously increasing attention from both scientific and industrial sectors in basic research projects. If apomixis is engineered into sexual crops in a controlled manner, its impact on agriculture will be broad and profound. In fact, apomixis will allow clonal seed production and thus enable efficient and consistent yields of high-quality seeds, fruits, and vegetables at lower costs. The development of apomixis technology is expected to have a revolutionary impact on agricultural and food production by reducing cost and breeding time, and avoiding the complications that are typical of sexual reproduction (e.g., incompatibility barriers) and vegetative propagation (e.g., viral transfer). However, the development of apomixis technology in agriculture requires a deeper knowledge of the mechanisms that regulate reproductive development in plants. This knowledge is a necessary prerequisite to understanding the genetic control of the apomictic process and its deviations from the sexual process. Our molecular understanding of apomixis will be greatly advanced when genes that are specifically or differentially expressed during embryo and embryo sac formation are discovered. In our review, we report the main findings on this subject by examining two approaches: i) analysis of the apomictic process in natural apomictic species to search for genes controlling apomixis and ii) analysis of gene mutations resembling apomixis or its components in species that normally reproduce sexually. In fact, our opinion is that a novel perspective on this old dilemma pertaining to the molecular control of apomixis can emerge from a cross-check among candidate genes in natural apomicts and a high-throughput analysis of sexual mutants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the greatest success stories in modern agriculture has been the tremendous yield increase achieved by coupling high-yield varieties with high-input agronomic systems, creating the so-called Green Revolution. Approximately one-third of the world’s seed supply comes from the commercial seed market, another one-third is provided by publicly funded institutions, and the seed saved by farmers accounts for the remainder.
Over the centuries, crop plants have followed the general pattern of introduction, selection, and hybridization. Crop introduction has been crucial for agriculture because many of the world’s crops are produced outside their region of domestication. Once introgressed, selection and breeding strategies have led to the development of new cultivars with improved yield and adaptation. Plant breeders are working to extend the Green Revolution by intensifying selection, developing more hybrid varieties in more crops, and increasing the range of plant functions through mutation and transgenic breeding. Hence, plant breeding will continue to play a crucial role in crop improvement because the needs are many, the techniques are expanding, the new genetic combinations are limitless, and the successes of the past illuminate the potential of the future. In outcrossing species, alleles disseminate in the offspring; thus, the optimal genotype is lost together with the desired trait. Exact copies of a superior genotype can be made via vegetative propagation; however, this technique is usually not applicable to annual crops such as maize, rice, and wheat. The fixation of a given genotype occurs naturally in species that exhibit an asexual type of seed production termed apomixis. This trait by itself is highly valuable for agriculture; however, despite many efforts, it has not been possible to introduce apomixis into modern domesticated crop species.
As a reproductive strategy for cloning plants via seeds, apomixis is a highly desirable trait in modern agriculture. In fact, apomixis results in offspring that are exact genetic replices of the female parent because embryos are derived from the parthenogenic development of apomeiotic egg cells (for reviews on apomixis, see Bicknell and Koltunow 2004; Ozias-Akins 2006; Albertini et al. 2010; Pupilli and Barcaccia 2012; Koltunow et al. 2013). From an evolutionary point of view, apomixis may be regarded as a consequence of sexual failure rather than as a recipe for clonal success (Silvertown 2008).
Introgression of apomixis from wild relatives into crop species and transformation of sexual genotypes into apomictically reproducing genotypes are long-held goals of plant breeding. Breeders believe that the introduction of apomixis into agronomically important crops will have revolutionary implications for agriculture. The potential benefits of harnessing apomixis are many and vary from full exploitation of heterosis by reseeding the best hybrids to clonal propagation of the superior genotypes in seed-propagated outcrossing crops. The impact of apomictic crops in agriculture would be massive in both developed and developing countries. Unfortunately, barring a few exceptions in some forage grasses and fruit trees, apomixis is not a common feature among crop species.
The fixation of hybrid vigor through apomixis is a desirable objective for breeders and farmers alike and is expected to have a revolutionary impact on food and agriculture production. The stabilization of heterozygous genotypes via apomixis would make breeding programs faster and cheaper (Fig. 1). The impact of apomictic crops in agriculture would be comparable to, or even greater than, the impact of the Green Revolution, especially in Third World countries (Vielle-Calzada et al. 1996; Pupilli and Barcaccia 2012). In fact, it has been estimated that the use of apomixis technology in the production of hybrid rice alone could provide benefits exceeding 1,800 million Euros per year (Spillane et al. 2004; Albertini et al. 2010). Apomixis technology could also provide benefits for clonally propagated crops. Clonal crop yields are limited by pathogens (mainly viral and endophytic), which accumulate over successive rounds of vegetative propagation and seriously limit the yield and exchange of germplasms between countries. The use of apomixis technology in these crops would provide the additional option and benefit of propagation via clonal seeds and thus generating disease-free material that can be more easily stored and transported. The use of apomictic seed as an alternative to vegetative propagules would provide similar benefits (e.g., lower costs and higher yields) over the current use of true seed of such crops. For example, apomixis technology could make true potato seeds a more attractive option for potato breeders and cultivators and would return benefits to growers of as much as 2.3 billion Euros per year (Spillane et al. 2004).
Comparison between conventional and apomixis-mediated methods for breeding F1 hybrid varieties. In traditional breeding, within a segregating population (e.g., F2 population) some genotypes are selected and after some generation of selfing followed by phenotypic selection, tested for their specific combining ability in order to be used as parental lines for the constitution of heterotic F1 hybrid seeds. The best performing inbred lines are selected, multiplied in isolated fields, and crossed in pairwise combinations to obtain uniform, vigorous, and high-yield F1 hybrids. This scheme, however, requires a series of actions: the two inbred lines must be kept pure and multiplied in separate fields. Then, to obtain the hybrid seed, it is necessary to establish a dedicated field where about one quarter of the plants is used as pollinator (i.e., pollen donor inbred) and on the remaining plants (i.e., seed parent inbred) the hybrid F1 seeds will be harvested. Farmers cannot re-use seeds collected from F1 hybrids as these seeds will give rise to highly variable populations because of genetic segregation and recombination. Using apomictic lines, however, the situation would be much simpler. Once superior inbred lines to be used as seed parent are selected, they can be crossed with clonal lines as pollen donors carrying the gene for apomixis, in order to obtain F1 hybrid seeds sharing a highly heterozygous genotype. From this moment on, each F1 hybrid variety can be maintained for several generations with permanently fixed heterosis
The development of apomixis technology in agriculture will require a deeper knowledge of the mechanisms regulating reproductive development in plants. Our molecular understanding of apomixis would be greatly increased if genes that are specifically or differentially expressed during the formation of the embryo and embryo sac could be identified. Over the last two decades, many scientists have speculated about the isolation of gene/s controlling key steps of the apomictic pathway, and many papers have postulated the production of engineered plants exhibiting apomictic-like phenotypes. In fact, none of the major crop plants have been bred for apomixis, and only some features of apomixis have been genetically engineered in model species. Consequently, even in the era of genomics, achieving an understanding of the genetic control and molecular regulation of apomixis appears much more complicated than expected. Large amounts of cytological and ecological information, along with genetic and molecular data, have been collected mainly from model species (i.e., Boechera holboellii, Hieracium spp., Hypericum perforatum, Paspalum spp., Poa pratensis, Ranunculus spp., and Taraxacum officinale) and have often been tested in Arabidopsis thaliana (Arabidopsis) to elucidate the mechanisms of apomeiosis, parthenogenesis, and apomixis. Several genes involved in the formation of unreduced embryo sacs and egg cells, in addition to genes responsible for the autonomous development of the embryo and endosperm, have been cloned and characterized; however, none of these genes are capable of miming the apomictic pathway as a whole in crop plants. Hence, after two decades of substantial studies conducted in several laboratories and model plants, the asexual reproductive strategy termed “gametophytic apomixis” by Nogler (1984) still appears to be an unsolved puzzle. As a result, seed companies have lost interest in this research, and it has been difficult to acquire funds for conducting research on apomixis.
Currently, novel views and original concepts are emerging from the fog, including a link between apomixis and gene-specific silencing mechanisms (likely based on chromatin remodeling factors or trans-acting and heterochromatic interfering RNAs involved in both transcriptional and post-transcriptional gene regulation) and the parallel between the Y chromosome and apomixis-bearing chromosomes from the most primitive to the most advanced in evolutionary terms (comparative genomic analyses revealed common features such as few recombination events, accumulation of transposable elements, and degeneration of genes). More recently, merging lines of evidence regarding the role of auxin in cell fate specification of the embryo sac and egg cell development have been reported in Arabidopsis.
Mechanisms of apomixis
Because apomictic reproduction entails the development of an embryo from a cell with a somatic chromosome number, there are several ways to produce embryos of apomictic origin. The simplest pathway avoids the production of an embryo sac, and the maternal embryo originates from one or more somatic cells of the ovule. Among the agriculturally important species, adventitious embryony (i.e., sporophytic apomixis) has been noted in mango (Mangifera indica), several Citrus species, and orchids. The most comprehensive treatise on adventitious embryony has been published by Naumova (1992).
When the maternal embryo originates from a diploid egg cell differentiated in an unreduced embryo sac, the apomictic pathway is referred to as gametophytic apomixis (Nogler 1984).
In gametophytic apomixis, the unreduced embryo sac may arise from a somatic nucellar cell that acquires the developmental program of a functional megaspore, a mechanism referred to as apospory. Alternatively, if the embryo sac forms from a megaspore mother cell with suppressed or modified meiosis, the pathway is referred to as diplospory. It is worth emphasizing that apomictic plants may or may not change meiosis itself, but in any case they do activate the gametic cell fate either in a somatic cell (apospory) or in an unreduced megaspore (diplospory) as surrogate for meiotic products (Albertini and Barcaccia 2007). Once 2n female gametophytes and gametes are formed (apomeiosis), they subsequently undergo embryogenesis autonomously without fertilization by a male gamete (somatic parthenogenesis). Endosperm formation may be fertilization-independent (autonomous endosperm) or may require fertilization (pseudogamous endosperm). Among others, apospory has been reported in Beta, Brachiaria, Cenchrus, Chloris, Compositae, Eriochloa, Heteropogon, Hieracium, Hyparrhenia, Hypericum, Panicum, Paspalum, Pennisetum, Poaceae, Ranunculus, Sorghum, Themeda, and Urochloa, whereas diplospory has been noted in Agropyrum, Allium, Antennaria, Boechera (formerly Arabis), Datura, Eragrostis, Erigeron, Eupatorium, Ixeris, Parthenium, Paspalum, Poa, Taraxacum, and Tripsacum (Table 1).
Inheritance of apomixis: genetic control and recombination potentials
In both aposporic and diplosporic species, robust models have been postulated and eventually validated for inspecting the genetic basis of apomixis and its components (i.e., apomeiosis and parthenogenesis). The inheritance fundamentals include a divergence in the number of genes, gene functions, and relationships among alleles, as well as dominance of apomixis over sexuality (Asker and Jerling 1992; Carman 1997; Savidan 2000; Grimanelli et al. 2001; Koltunow and Grossniklaus 2003). Genetic analysis in several species has consistently demonstrated that a simple inheritance system involving a few Mendelian genes controls the expression of apomixis or its components. In contrast, molecular and cytogenetic analyses of chromosomal region(s) carrying the determinants of apomixis in several species have revealed a complex genetic control mechanism that is likely based on a system of polygenes in addition to mechanisms involving a lack of recombination, trans-acting elements for gamete elimination, supernumerary chromatin structures, and DNA rearrangements (see a review by Pupilli and Barcaccia 2012 and references therein).
Currently, gametophytic apomixis is thought to rely on three genetically independent Mendelian loci, each exerting control over a key developmental component, including apomeiotic megaspores, parthenogenic unreduced egg cells, and modified endosperms (Grossniklaus et al. 2001a; Koltunow and Grossniklaus 2003; Bicknell and Koltunow 2004; Vijverberg and van Dijk 2007; see also a review by Albertini et al. 2010 and references therein). A single regulatory gene was originally proposed as being sufficient for promoting apomixis (Peacock 1992). Although simple genetic control seems to support this hypothesis, molecular evidence suggests that a more complex inheritance system directs the entire process of apomixis. In some species, linkage groups typically transmitted with apomixis contain large blocks of sequence that lack genetic recombination between molecular markers, leading to speculation that adapted gene complexes within supernumerary chromatin might be required for the expression of apomixis (Ozias-Akins et al. 1998; Roche et al. 2001; Akiyama et al. 2004). A close relationship between apomictic mechanisms and heterochromatic regions of the genome that are rich in retrotransposons has raised the intriguing possibility that DNA structure and/or RNA interference could play a role in regulating the expression of apomixis-related genes (Pupilli and Barcaccia 2012). A well-characterized class of small regulatory RNAs that are widespread in eukaryotes appears to regulate gamete function and fertilization in plants by altering gene expression through post-transcriptional gene silencing, translational inhibition, and heterochromatin modification (Ron et al. 2010).
Increasing experimental evidence suggests that genetic recombination can either be suppressed or allowed in chromosomal regions surrounding the master locus for apomixis depending on the evolutionary pathway of the genome. This is a phenomenon that has been well documented for plant chromosomes carrying sex-determining genes (Vyskot and Hobza 2004). From an evolutionary point of view, we expect that relatively young and simple genetic systems of apomixis determination should include a narrow euchromatic region where genetic recombination between apomeiosis and parthenogenesis loci, and their linked genes, is possible (Table 1), as in Poa (Barcaccia et al. 2000; Albertini et al. 2001), Taraxacum (van Dijk and Bakx-Schotman 2004), Hypericum (Schallau et al. 2010), Erigeron (Noyes and Rieseberg 2000), Hieracium (Catanach et al. 2006), and Panicum maximum (Kaushal et al. 2008). In contrast, a degenerate heterochromatic block carrying apomixis factors should represent evolutionarily advanced genetic systems of apomixis determination with large non-recombining regions surrounding the apomixis locus (Table 1) as in Pennisetum/Cenchrus (Ozias-Akins et al. 1998; Roche et al. 1999), Brachiaria (Pessino et al. 1998), Paspalum (Labombarda et al. 2002; Stein et al. 2007; Podio et al. 2012), and Tripsacum (Grimanelli et al. 1998). Just recently, Conner et al. (2013) found recombination between apospory and parthenogenesis loci in C. ciliaris (Table 1). Based on the experimental data collected thus far, Pupilli and Barcaccia (2012) have recently hypothesized that a relatively simple genetic system controls apomixis in terms of the number of genes involved in the expression of its components (i.e., genes controlling apomeiosis and parthenogenesis and eventually autonomous endosperm development). However, elements within the chromosome block carrying the apomixis genes (e.g., transposable elements, repetitive elements, and pseudogenes) make it a complex genetic system, with loci that vary from elementary and primitive to evolutionarily advanced. According to recent findings, the first type reflects a chromosome pair showing tightly linked genetic determinants for apomixis in a narrow euchromatic region where genetic recombination is not suppressed, whereas the other type includes a chromosome pair that possesses a degenerate gene block with a large non-recombining region surrounding the apomixis locus (for details see Pupilli and Barcaccia 2012).
Searching for genes controlling apomixis in natural apomicts
Although many years of descriptive studies have provided a solid documentation of the types of apomictic processes that occur in a wide variety of plant species, molecular studies aimed at understanding the basis of apomixis have failed to adequately elucidate its central mystery, partly because the majority of apomicts do not constitute agriculturally important crops and, with a few exceptions (e.g., Tripsacum and maize), do not have agriculturally important relatives (Bicknell and Koltunow 2004; Albertini et al. 2010). An early theory regarding genetic control of apomixis proposed that the trait is regulated by “a delicate gene balance” (Muntzing 1940) of recessive genes and that this balance might be disturbed after crosses. Currently, basic inheritance is usually thought to depend on a single master regulatory gene or a few dominant key genes, which allow a megaspore mother cell or a somatic nucellar cell to form an embryo sac without meiotic reduction and an embryo to develop from an unreduced egg cell without fertilization (Asker and Jerling 1992; Koltunow et al. 1995; Savidan 2000; Grossniklaus et al. 2001b). Once apomictic genes initiate embryo development and the initial cell forms and divides, the genes controlling embryo cell formation and patterning are most likely the same as those required for sexual embryo development. Whether the products of apomictic genes are proteins that are not produced in sexually reproducing plants (i.e., gain of function) or proteins that normally function to initiate events in sexual reproduction but have become altered with respect to their activity or spatial and temporal distribution during development (i.e., loss of function) is still not well understood. Currently, a number of researchers support the hypothesis that zygotic embryogenesis and apomictic parthenogenesis follow similar pathways during embryo and seed production (Bicknell and Koltunow 2004; Albertini et al. 2004; Sharbel et al. 2010). Specific genes are activated, modulated, or silenced in the primary steps of plant reproduction to ensure that functioning embryo sacs develop from meiotic spores and/or apomictic cells. Because additional genes may be specifically or differentially expressed in sexually versus apomictically reproducing plants, and these genes may operate during embryo development, we would be better equipped to understand apomixis if the genes responsible for controlling specific and differential expression during embryo and embryo sac formation could be identified.
Some scientists believe that apomixis is controlled by specific genes encoding new proteins with a novel initiating function not observed in sexually reproducing plants, and these scientists have performed experiments based on either differential display or subtractive hybridization of a single flower stage. This led to the identification of a number of candidate genes. For example, in Brachiaria species, among the 12 candidates isolated by Leblanc et al. (1997), only two proved to be specifically expressed in mature ovaries containing unreduced (aposporic) embryo sacs. Instead of genes specifically expressed either in apomictic or sexual genotypes, Rodrigues et al. (2003) searched for candidates expressed in both genotypes of the same species and identified 11 genes that were differentially expressed between apomictic and sexual genotypes. In Paspalum notatum, three distinct gene transcripts showed differential expression between apomictic and sexual F1 individuals after apospory initiation in flowers (Pessino et al. 2001). An additional 65 genes that were differentially expressed between apomictic and sexual genotypes at the meiotic stage were identified by Laspina et al. (2008). A large subset of these candidates mapped in silico to a genomic region on rice chromosome 2 that was previously associated with apospory (Pessino et al. 1998; Pupilli et al. 2004), and one of these genes showed high similarity to lorelei, a gene associated with male gamete delivery to the egg cell in Arabidopsis (Felitti et al. 2011).
Candidate genes specifically expressed in either apomictic or sexual ovules were also identified in P. maximum (Chen et al. 1999, 2005; Yamada-Akiyama et al. 2009) and Pennisetum ciliare (Vielle-Calzada et al. 1996). Additional genes differentially expressed between apomictic and sexual samples were isolated in P. maximum (Yamada-Akiyama et al. 2009), P. ciliare (Vielle-Calzada et al. 1996; Singh et al. 2007), Hieracium pilosella (Guerin et al. 2000), and Eragrostis curvula (Cervigni et al. 2008; Selva et al. 2012).
Although a large number of candidate genes exhibiting differences in spatial and temporal expression levels and patterns have been identified with these approaches, both their function and their involvement in the control of apomixis remain largely speculative (Ozias-Akins 2006). Moreover, although negative in context, these studies added support to the second theory, which proposes that apomixis is controlled by proteins that normally function to initiate events in sexual reproduction but may be altered with respect to their activity or spatial and temporal distribution during development (Bicknell and Koltunow 2004). One of the first supporters of this hypothesis was Carman (1997), who suggested that apomixis is a result of the deregulation of sex-related genes with respect to spatial and temporal expression as a consequence of their heterochronic expression due to hybridization (Singh et al. 2007; Sharbel et al. 2009; reviewed by Albertini et al. 2010). Research carried out in species that reproduce through distinct pathways seemed to confirm that apomixis relies upon either spatial or temporal misexpression of genes acting during female sexual reproduction (Grimanelli et al. 2003; Tucker et al. 2003; Albertini et al. 2004; Curtis and Grossniklaus 2007; Sharbel et al. 2009). Attempts to isolate asynchronously regulated genes were carried out by comparing the transcriptional profiles of apomictic and sexual ovules over several developmental stages in several species because careful staging was thought to be critical for the interpretation of the results, particularly if misexpression, rather than unique expression, was responsible for the switch in reproduction mode (Ozias-Akins 2006). Recently, careful staging of ovary development has led to the identification of differentially expressed transcripts in P. pratensis (Albertini et al. 2004, 2005; Marconi et al. 2013), B. holboellii (Sharbel et al. 2009, 2010), Paspalum simplex (Polegri et al. 2010), and H. perforatum (Galla and Barcaccia 2012; Galla et al., personal communication).
In P. pratensis, Albertini et al. (2004) isolated as many as 179 fragments that were differentially expressed between apomictic and sexual genotypes. Importantly, most of the transcripts were not specifically associated with apomictic or sexual genotypes; instead, expression was differentially modulated or quantitatively different (Albertini et al. 2004, 2005), supporting the hypothesis that apomixis may result from a deregulated sexual pathway. In particular, PpSERK and APOSTART (Table 1) were characterized in detail (Albertini et al. 2005) and were thought to be involved in cell-to-cell signaling and hormone trafficking. The authors proposed that PpSERK activation in nucellar cells of apomictic genotypes is the switch that triggers embryo sac development and could redirect signaling gene products to compartments other than their typical ones. The SERK-mediated signaling pathway may interact with the auxin/hormonal pathway controlled by APOSTART. Indeed, based on its temporal and spatial expression patterns, APOSTART is potentially associated with apomixis, and its transcripts are detectable specifically in aposporic initials and embryo sacs. Additionally, gene expression studies of gene members revealed a delay of APOSTART6 expression in apomictic and parthenogenic P. prantensis genotypes (Fig. 2, panels a–e), supporting an involvement of this allele in parthenogenesis (Marconi et al. 2013). Overall, the accumulated data suggest that APOSTART may be related to programmed cell death (PCD) that is involved in the non-functional megaspore and nucellar cell degeneration events that permit enlargement of maturing embryo sacs. Functional characterization of the Arabidopsis APOSTART1 gene (AtAPO1) showed that it is expressed in mature embryo sacs and developing embryos. APOSTART1/APOSTART2 double mutants seem to confirm an involvement of this gene in embryo/seed development (Albertini et al. unpublished data).
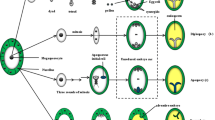
Expression data related to candidate genes for apomixis. Gene expression patterns and levels of APOSTART6 in P. pratensis and ARIADNE7 in H. perforatum as assessed by in situ hybridization and real-time RT-PCR analysis. a–d APOSTART6 expression patterns in longitudinal sections of P. pratensis ovaries: signal is present in one or more nucellar cells (arrow) within the ovule of apomictic genotypes (a) and in the megaspore mother cell in sexual genotypes (data not shown, for details see Albertini et al. 2005). Signal is then present during embryo sac development (b, c) and embryo development (d). e Expression patterns and level of transcripts encoded by APOSTART6 in apomictic (dark blue), sexual (red), and parthenogenic recombinant (light blue) genotypes of P. pratensis (for details see Marconi et al. 2013). Delay of expression in apomictic and parthenogenetic genotypes suggests an involvement of APOSTART6 in parthenogenesis. f, g Longitudinal sections of H. perforatum ovules at the stage of female meiosis showing hybridization signals of ARIADNE7 transcripts (arrows) in correspondence with nucellar tissues next to megaspores; h negative control (courtesy of Giulio Galla, University of Padova). i Expression levels of the ARIADNE7 transcripts in young buds, anthers, pistils, and sepals and petals: this gene was found preferentially expressed in pistils and young buds of apomictic genotypes. Specificity of expression domain in apomictic and aposporic genotypes suggests an involvement of ARIADNE7 in apospory
More recently, Sharbel et al. (2009, 2010) utilized a high-throughput differential display approach to study naturally occurring quantitative variations in gene expression between ovules of apomictic and sexual B. holboellii genotypes and identified 543 genes exhibiting a developmental shift in expression between the sexual and apomeiotic ovules. A carefully devised experiment was also undertaken by Polegri et al. (2010), leading to the identification of a set of genes in P. simplex with stage- and/or phenotype-specific expression. In particular, a class of alleles that showed a characteristic specificity of expression was fully linked to apomixis based on mapping data.
In H. perforatum, a differential display analysis of sporogenesis and gametogenesis led to the isolation of several transcripts specifically expressed in the pistils of a highly apomictic ecotype, including an EST showing similarity to a gene coding for an ATPase RNA helicase responsible for an embryo defective phenotype in Arabidopsis (MEE29, maternal effect embryo). This gene, termed HpMEE29-like, was differentially expressed between aposporic and meiotic H. perforatum plants (Barcaccia et al. 2007). More recently, Galla and Barcaccia (2012) adopted high-throughput 454 technology to sequence the entire Hypericum flower transcriptome using single verticils collected from apomictic and sexual genotypes. Computational procedures were used to assemble and annotate more than 25,000 transcripts exclusively from anthers and carpels at different developmental stages. Galla et al. (2013) identified dozens of genes related to sporogenesis and gametogenesis, with particular reference to the formation of embryo sacs, embryos, and seeds. Interestingly, some of the transcripts showed sequence homology with candidate genes for apomixis that were cloned in apomictic species and mutants, including components of the HAPPY locus (Schallau et al. 2010). Notably, many small transcripts of genes specifically expressed in apomictic genotypes exhibited high similarity to microRNA precursors that target specific transcription factors. In particular, the most represented and conserved families were miR156, miR166, miR390, miR394, miR396, and miR414, which have dozens of potential target genes with a wide range of molecular functions including metabolism, response to stress, flower development, and plant reproduction Galla et al. (2013).
Different approaches were chosen in Pennisetum squamulatum (Conner et al. 2008) and H. perforatum (Schallau et al. 2010). In P. squamulatum, by sequencing an apospory-specific genomic region (ASGR), Conner et al. (2008) isolated a gene sharing similarity with BABY BOOM (BBM), which was named ASGR–BBM (Table 1). This gene encodes a protein containing two AP2 domains that are 96 % similar to the AP2 regions of BnBBM (outside the AP2 domains, the similarity of ASGR-BBM to BnBBM declines significantly to 35 and 27 % in the upstream and downstream regions, respectively). In H. perforatum, Schallau et al. (2010) screened genomic clones using an apospory-linked SCAR marker as probe and identified a 142 kb BAC clone containing a gene homologous to Arabidopsis ARIADNE7 (ARI7, Table 1), which is annotated as a nucleic acid binding protein. In particular, both aposporic- and sexual-specific HpARI7 alleles were found co-expressed in the pistils at different developmental stages, whereas the gene product of the apomictic allele was specifically expressed in pistils of the apomictically reproducing individuals (Schallau et al. 2010). More recently, Galla et al. (personal communication) demonstrated that HpARI7 gene is preferentially expressed in pistils at different developmental stages and that HpARI7 transcripts are specifically detectable in nucellar tissues of the ovule next to megaspores in apomictic H. perforatum genotypes (Fig. 2, panels f–i). Specificity of the expression domain in apomictic and aposporic genotypes suggests an involvement of this gene belonging to the HAPPY locus in apospory.
Functional analysis of genes miming apomixis in sexual model plants
If it is true that apomixis is a consequence of sexual failure, rather than a means for clonal success, from an evolutionary point of view (Silvertown 2008), it is also true that apomixis, as a biological process of seed formation, represents an altered form of sexuality rather than a new developmental program (Koltunow and Grossniklaus 2003). Carman (1997) hypothesized that apomixis is a result of the spatial and temporal deregulation of sex-related gene expression as a consequence of heterochronic expression due to hybridization. This hypothesis was confirmed experimentally by cytological observations in Tripsacum (Grimanelli et al. 2003) and more recently by transcriptome analyses in Boechera (Sharbel et al. 2009, 2010). Additionally, Tucker et al. (2003) provided evidence that marker genes related to embryo sac development are similarly expressed in sexual and apomictic Hieracium genotypes.
The idea that apomixis is an altered form of sexuality that results from temporal and spatial alterations in the action of the sexual program suggests that a synthetic apomixis system using variant alleles of genes isolated from sexual model species, such as Arabidopsis, can be developed (Chaudhury and Peacock 1993). By identifying and combining the genes involved in apomixis, the assembly of an asexual system of seed formation in a sexual plant should be possible. In particular, some of the genes isolated and characterized from sexual species may have roles in apomixis (Table 2). For example, the recent findings regarding the molecular mechanisms controlling embryo sac development, fertilization, and endosperm development may be useful for determining genetic links with apomeiosis, parthenogenesis, and autonomous or pseudogamous endosperm development. It is believed that basic structural and functional analyses of these candidate genes are crucial for engineering apomixis in sexual crops.
Genes that are expressed during embryo sac development, and thus putatively involved in differentiating sexual from apomictic pathways, are of particular interest. An embryo sac develops through megaspore mother cell (MMC) differentiation, meiosis, determination of the functional megaspore, and embryo and endosperm development. Because meiosis is either completely bypassed (apospory) or extremely altered (diplospory) in apomixis, genes related to female sporogenesis are thought to be more specifically involved in diplosporic apomixis, and genes involved in embryo sac cell identity are presumably crucial for aposporic apomixis. Overall, genes associated with female gametogenesis and egg cell development are likely shared between sexual and apomictic pathways (for details on these aspects see reviews by Tucker et al. 2003; Koltunow and Grossniklaus 2003; Ozias-Akins and van Dijk 2007; Tucker and Koltunow 2009; Albertini et al. 2010; Dwivedi et al. 2010).
Regarding the determination of the megasporocyte, an analysis of mutations affecting MMC differentiation may be crucial for understanding the specification of aposporic initials during female sporogenesis. The Arabidopsis mutant sporocyteless/nozzle (spl) is unable to develop a functional MMC and shows defects in nucellar cell identity (Schiefthaler et al. 1999; Sieber et al. 2004). Moreover, mutations in the Arabidopsis gene WUSCHEL (WUS), a regulator of stem cell identity in the shoot apical meristem, also result in defects in MMC specification (Gross-Hardt et al. 2002). WUS acts by modulating the expression of WINDHOSE1 (WIH1) and WINDHOSE2 (WIH2), which control female sporogenesis in conjunction with TORNADO2 (Lieber et al. 2011). Additionally, the MULTIPLE ARCHESPORIAL CELLS1 (MAC1) gene of maize (Sheridan et al. 1996, 1999) and the MULTIPLE SPOROCYTES1 (MSP1) and TAPETUM DETERMINANT LIKE1A (TDL1A) genes in rice (Nonomura et al. 2003; Zhao et al. 2008) are required for the production of only one megasporocyte per single ovule, indicating that these genes are involved in MMC determination. Loss of gene function results in multiple MMCs, indicating that these genes negatively regulate the sporogenous cell fate in the ovule. Zhao et al. (2002) identified the ortholog of MSP1 in Arabidopsis (EXTRA SPOROGENOUS CELLS7 EXCESS MICROSPOROCYTES1, EXS7EMS1); however, its role in regulating MMC number is not known. MSP1 encodes a leucine-rich repeat-like kinase (LRR–RLK) (Nonomura et al. 2003) that has been identified in plants as a transmembrane protein involved in a complex array of signaling pathways related to cell differentiation and developmental events (Diévart and Clark 2004).
In Arabidopsis, an alternative route for restricting the specification of embryo sac precursors has been proposed through the action of ARGONAUTE (AGO) genes (Olmedo-Monfil et al. 2010; Tucker et al. 2012). AGO proteins are known to be involved in post-transcriptional gene silencing mediated by short RNAs (either microRNAs or short interfering RNAs) (Baumberger and Baulcombe 2005). Small RNAs (sRNAs) have recently been studied in different model systems, and it is now known that mutations in the molecular pathways that generate sRNAs may dramatically affect fertility (Van Ex et al. 2011; Tucker et al. 2012). Previous research has demonstrated that strong mutant alleles of genes involved in the formation and activity of miRNAs, such as AGO1, DCL1, HEN1, and HYL1, disrupt reproductive development (reviewed by Van Ex et al. 2011). However, interpreting these phenotypes is frequently difficult because such mutations have ectopic effects and influence different aspects of plant development (Axtell 2013). Two members of the ARGONAUTE protein family, AGO5 and AGO9, which are involved in the regulatory pathway of sRNAs in plants, have been associated with cell specification and embryo sac development (Olmedo-Monfil et al. 2010; Tucker et al. 2012). An AGO5 ortholog in rice was reported to be essential for the progression of pre-meiotic mitosis and meiosis (Nonomura et al. 2007), and the production of viable gametes without meiosis was reported in maize lacking the ortholog of AGO9 (Singh et al. 2011). Some of the genes isolated and characterized from sexual species may play roles in the framework of apomixis, and it is possible that sRNAs act by silencing master genes directly involved in differentiating apomictic from sexual pathways.
Particular Arabidopsis mutants have revealed that gametogenesis can be uncoupled from meiosis. For example, loss of certain ARGONAUTE (i.e., AGO9) genes and other genes in the small RNA pathway, such as RNA-DEPENDENT RNA POLYMERASE6 (RDR6) and SUPPRESSOR OF GENE SILENCING3 (SGS3), resulted in loss of restriction in gametic cell identity and fate in the ovule and gain of expression in multiple somatic initials in the nucellar tissue that can differentiate into gametic cells without undergoing meiosis and can initiate female gametogenesis through the activation of TEs (Olmedo-Monfil et al. 2010). Notably, these TEs are normally silenced in both developing and fully differentiated Arabidopsis ovules (Slotkin et al. 2009). AGO9 silences TEs in the embryo sac, and hence, its function resembles that of the PIWI (P-element-induced wimpy testis in Drosophila) regulatory proteins responsible for maintaining incomplete differentiation in stem cells and preserving stable cell division rates in the germ line lineage of invertebrates and mammals (Klattenhoff and Theurkauf 2008). A phenotype similar to that generated by AGO9 loss-of-function alleles has also been observed in maize by repressing DMT103 and DMT102, which are homologs of Arabidopsis DOMAINS REARRANGED METHYLTRANSFERASE (DRM2) and CHROMOMETHYLASE 3 (CMT3) genes, respectively (Garcia-Aguilar et al. 2010). These genes are involved in new DNA methylation in DRM2 sequences and in the retention of DNA methylation in non-CG sites in Arabidopsis (Cao and Jacobsen 2002). Although the function of these genes is unknown in maize, they could play a role in ensuring that a single embryo sac develops within each ovule. This mechanism could be epigenetically regulated through DNA methylation (Pillot et al. 2010a, b).
Evidence regarding the role of auxin in the cell fate specification of embryo sac development has recently been obtained. In Arabidopsis, two YUCCA (YUC) genes encoding proteins that are crucial for local auxin biosynthesis were shown to be expressed in the ovule, which is consistent with the role of auxin as a cell fate determinant (Pagnussat et al. 2009). Owing to the high concentrations of auxin detected in the distal tip of the nucellus at early stages of ovule development (Pagnussat et al. 2009) and because SPOROCYTELESS/NOZZLE (SPL) was demonstrated to repress the expression of YUCCA genes (Li et al. 2008), it is likely that auxin plays a key role in the cell specification machinery that regulates differentiation of the MMC and/or maintains the undifferentiated state of nucellar cells once the MMC is formed.
Proper MMC formation involves two distinct pathways: the first involves a commitment to the specification of a MMC from a somatic cell (i.e., cellular identity determination), and the second involves a commitment to the differentiation of an MMC from a single cell (i.e., cellular type specialization). Knowledge of key genes involved in both pathways should provide the molecular tools necessary for developing an artificial apomictic system in sexual plants.
In Arabidopsis, as in most angiosperms, the MMC undergoes regular meiosis and gives rise to a tetrad of haploid megaspores, of which three usually degenerate and one becomes the functional megaspore. Several loss-of-function phenotypes related to megasporogenesis were recently discovered in Arabidopsis and monocots such as rice and maize, with each showing some features of diplospory. For example, in the rice mutant meiosis arrested at leptotene1 (mel1), the MMCs arrested megasporogenesis at pre-meiotic or eventually at meiotic stages (Nonomura et al. 2007). The gene controlling the mel1 phenotype belongs to the ARGONAUTE (AGO) gene family, which is involved in several developmental processes in plants via the action of sRNAs (Vaucheret 2008). Another gene that is critical for proper megasporogenesis in Arabidopsis is DYAD/SWITCH1 (SWI1), which is responsible for sister chromatid cohesion and centromere organization at meiosis. An allelic variant of this gene, dyad, proved to be responsible for the production of few unreduced egg cells (Ravi et al. 2008). Fully penetrant diplospory-like phenotypes were induced in Arabidopsis by replacing meiosis with mitosis in MiMe genotypes (d’Erfurth et al. 2009, 2010). In particular, these genotypes combine mutations in the two genes SPO11-1, which prevents chromosome pairing and recombination, and REC8 (also known as SYN1), which modifies chromatid segregation along with OSD1 (MiMe-1 mutant; d’Erfurth et al. 2009) or CYCA1-2/TAM (MiMe-2 mutant; d’Erfurth et al. 2010). Similar to dyad, these mutants were shown to form unreduced egg cells because they did not undergo a second meiotic division. Singh et al. (2011) uncovered a maize mutant, Dominant non-reduction 4 (Dnr4), that manifested defects in chromatin condensation during meiosis with subsequent failure of chromosome complement segregation, suggesting that this mutation affects the function of chromatin remodeling factors. After substitution of a meiosis division for a mitosis-like division, these mutants formed functional unreduced egg cells that exhibited phenotypes resembling diplospory, which is similar to what was observed in the maize elongate (el1) mutant (Rhoades and Dempsey 1966) and the natural apomict Tripsacum (Grimanelli et al. 2003).
In maize, the Dnr4 mutant phenotype arises due to lesions in the AGO104 gene, which is the ortholog of Arabidopsis AGO9. Together with MEL1, these genes belong to the ARGONAUTE gene family. The abnormal patterns of cell specification mediated by the lack of AGO9 and AGO104 are reminiscent of aposporic and diplosporic apomixis, respectively, suggesting that these natural apomeiotic variants might rely on similar regulatory pathways although they are characterized by fundamental differences.
Concerning the selection of the functional megaspore, in Arabidopsis as in most of Angiosperms, the MMC undergoes regular meiosis and gives rise to a linear tetrad of haploid megaspores. Three of these megaspores (usually those located in the micropylar area) degenerate, and only one becomes the functional megaspore that undergoes meiosis. A distinctive process that occurs in aposporic species (e.g., Paspalum spp., H. perforatum) is the degeneration of all four megasporocytes while the already advanced aposporic embryo sacs develop further; in diplosporic species, unreduced megaspores originate via restitutional meiosis (i.e., Taraxacum-type, which is also widespread in Boechera spp.) or via a complete bypass of meiosis (i.e., Antennaria-type, also common in Tripsacum dactyloides).
In diplospory, abnormal meiosis is largely asynaptic because of the absence of pairing between homologous chromosomes and therefore results in a restitution nucleus in the first division and the formation of a dyad of unreduced megaspores (for a review, see Albertini et al. 2010). In addition to diplospory, several molecular mechanisms associated with apospory may also explain the specificity of megasporocyte degeneration, including position-dependent degeneration, cell polarization changes, and PCD (reviewed by Pupilli and Barcaccia 2012).
The easiest explanation for the occurrence of apomeiosis could be the fact that the chalazal megaspore has preferential access to nutritional elements or regulatory factors because it is closest to the maternal tissues. In aposporic ovules, the hierarchical position of the linear tetrad of megaspores could be perturbed by the growth of multiple embryo sacs that compete for nutrients and growth factors (Pupilli and Barcaccia 2012). The question remains, what happens in diplosporic ovules when meiosis is altered or bypassed? Molecular studies in Arabidopsis have suggested that the degeneration and death of the micropylar megaspore and the identity determination of the functional megaspore may be controlled by a positional signaling pathway (Yang and Sundaresan 2000) and/or a polarity effect, as manifested by the distribution pattern of organelles and microtubules of the cytoskeleton during megasporogenesis (Bajon et al. 1999).
Among the characteristic features of cells undergoing PCD, DNA degradation and changes in Ca2+ accumulation dynamics have been observed in degenerating megaspores (as reviewed by Tucker and Koltunow 2009). In lettuce, differences between Ca2+ accumulation pathways in the degenerating micropylar megaspore and the chalazal megaspore were observed, and the latter retained a significantly higher concentration of calcium (Qiu et al. 2008). Some members of the MPS-ONE-BINDER (MOB1) gene family in diplosporic mutants of Medicago sativa were specifically expressed in degenerating megaspores of normal ovules and in enlarged megaspore mother cells and embryo sacs of apomeiotic ovules (Citterio et al. 2005, 2006). MOB1 gene products were also found in microspore tetrads at the beginning of pollen development and in the tapetum cells of anthers undergoing PCD to allow pollen dispersal at maturity. Overall, the results suggest that MOB1 genes can play a key role in the reproductive pathway in plants. MOB proteins are involved in cell cycle progression and PCD in Drosophila and mammals (Hirabayashi et al. 2008; Vitulo et al. 2007).
In Arabidopsis and some apomictic species, one factor putatively associated with the selective degeneration of meiotic megaspores is regulated by callose boundaries and plates that can accumulate at one pole or around the MMC and in the transverse cell walls between the functional megaspore and its degenerated sister megaspores (Olmedo-Monfil et al. 2010); these boundaries and plates can also form next to the unreduced megaspore of apomeiotic dyads in Medicago diplosporic mutants (Albertini and Barcaccia 2007). Another factor is the ANTIKEVORKIAN (AKV) gene that, when mutated, allowed for the survival of all four megaspores in 10 % of Arabidopsis ovules (Yang and Sundaresan 2000). However, in Paspalum and other aposporic grasses, complete suppression of the sexual pathways does not always occur (Hojsgaard et al. 2008, 2013). As a consequence, different degeneration patterns of megaspores in apomictic and sexual lineages cannot be governed by specific apomixis-related genes. This process is more likely to be pleiotropically affected by a gene cascade triggering the apomictic pathway; alternatively, the sexual megaspores could be mechanically disrupted by the overgrowing aposporic embryo sacs.
Another crucial step in apomictic reproduction is the initiation and autonomous development of the embryo combined with the formation of the endosperm. Parthenogenesis, as a fertilization-independent form of embryo development, is a key component of aposporic embryo development, and it is the only feature that aposporic development has in common with diplosporic embryo development. Parthenogenesis has been widely observed in nature; however, in sexual plants, it generally occurs at a low rate in haploid egg cells (Lacadena 1974). A general rule in zygotic development is that the activation and consequent initiation of embryogenesis is trigged by the fertilization of an egg cell. In animals, egg cell fertilization induces an increase in Ca2+ levels at the site of sperm cell entry that propagates throughout the egg cell, inducing a downstream cascade of signals required to initiate embryogenesis (Miyazaki and Ito 2006). In plants, an increase in intracellular Ca2+ was observed after egg cell fertilization (Antoine et al. 2000); however, this was not sufficient to trigger parthenogenesis (Curtis and Grossniklaus 2008).
Parallel mutant screens for apomixis enabled the identification of genes controlling the fertilization-independent initiation of seed development in Arabidopsis. These genes, termed FERTILIZATION-INDEPENDENT SEEDS (FIS) genes, encode protein members of the Polycomb-related complex (Koltunow and Grossniklaus 2003). The fis mutants are known to initiate endosperm development without fertilization to varying extents. However, the frequency of embryo initiation by division of an egg cell is either low or does not occur at all in most fis mutants.
Several natural mutations are known to induce the parthenogenic development of embryos in plants when the zygote is forced to begin its growth in a haploid environment. The “Salmon” system in wheat produces high numbers of haploid parthenogenic embryos (Matzk 1996), and the haploid inducer (hap) mutant of barley is associated with parthenogenesis (Hagberg and Hagberg 1980). More recently, it was shown that parthenogenic embryos can be generated at a relatively high frequency in transgenic lines of Arabidopsis expressing a modified centromere-specific histone CENH3 protein (Ravi and Chan 2010). However, the question of whether these findings could be ascribable to apomictic features in wild apomicts is unclear because these mutations are related to the haploid condition of the zygote. In Arabidopsis, the ubiquitous overexpression of several transcription factors induces ectopic formation of embryo-like structures. LEAFY COTYLEDON (LEC1 and LEC2) genes induce the expression of embryo-specific genes and trigger the development of embryo-like structures (Stone et al. 2001). PICKLE (PKL) is an upstream regulator of LEC genes that acts as repressor of embryogenesis (Henderson et al. 2004), and BABY BOOM (BBM) is an AP2-domain transcription factor that when overexpressed leads to the development of embryos and cotyledons from vegetative tissues (Boutilier et al. 2002). Moreover, the homeodomain protein encoded by WUSCHEL (WUS) is involved in promoting the vegetative to embryogenic transition and/or maintaining the identity of embryonic stem cells (Zuo et al. 2002). However, no experimental evidence supports similar functions for these genes in the egg cell or zygote. Of the genes proven to promote somatic embryogenesis in vegetative tissues, only the Arabidopsis ortholog of carrot SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) was expressed during gametogenesis and early zygotic embryogenesis (Hecht et al. 2001; Albertini et al. 2005). However, a clear phenotype could not be observed when the SERK gene was ectopically expressed in Arabidopsis (Hecht et al. 2001).
Currently, it is well known that the female gametophyte controls embryo and/or endosperm development at two different levels: (1) repression of embryo/endosperm development in the absence of fertilization through imprinting and (2) expression of factors that are required after fertilization. FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) (Ohad et al. 1999), MEDEA (MEA) (Grossniklaus et al. 1998), and FERTILIZATION-INDEPENDENT SEED2 (FIS2) (Chaudhury et al. 1997) repress endosperm development in the absence of fertilization. Mutations in another member of this group, MULTICOPY SUPPRESSOR OF IRA (MSI1), induce the formation of rudimentary parthenogenic embryos from growing and dividing egg cells and lead to the repressive FIS gene phenotype. Nevertheless, these parthenogenic embryos abort at an early stage (Guitton and Berger 2005). All these genes demonstrate homology with the Polycomb group (PcG) of genes that are involved in repressing development in Drosophila through chromatin remodeling mechanisms (Schwartz and Pirrotta 2007). The FIE/FIS2/MEA complex (FIS complex) acts by repressing the transcription of target genes involved directly in endosperm development. One of these genes, PHERES1 (PHE1), encodes a MADS domain-containing protein. MEA and FIE proteins interact directly with the PHE1 promoter (Köhler et al. 2003). All fis mutations show aberrant embryo and endosperm development if fertilized and exhibit autonomous endosperm development if unfertilized; however, the seeds abort irrespective of the paternal contribution.
Imprinting, or parent-specific expression of genes, is a mechanism by which the embryo sac controls the early stages of seed development. MEA was the first documented example of an imprinted gene involved in seed formation (Grossniklaus et al. 1998) in which the paternal allele is silenced and the maternal allele is expressed in the central cell. Both parental alleles are then silenced, and the modifications are removed in the central cell by a protein encoded by DEMETER (DME), a DNA glycosylase/lyase related to the DNA repair protein family (Choi et al. 2002). All genes of the FIS complex act through an epigenetically regulated mechanism to maintain the quiescent status of the central cell in the absence of fertilization. MULTICOPY SUPPRESSOR OF IRA (MSI1), another PcG gene product, participates together with its interacting cell cycle control protein RETINOBLASTOMA-RELATED (RBR) in the molecular machinery that represses autonomous endosperm development (Ebel et al. 2004). The MSI1/RBR complex regulates the methylation state of DNA during gametogenesis by repressing the transcription of a DNA methylase that normally controls methylation during embryo sac formation in Arabidopsis (Johnston et al. 2008; Jullien et al. 2008).
In plants, two main strategies have been adopted to demethylate DNA during gamete formation in ovules. One of these strategies, mediated by the MEA/DME system, is specific to the central cell, whereas the other strategy, controlled by the MSI1/RBR complex, is genomewide, affecting both the egg cell and the central cell (Sundaresan and Alandete-Saez 2010). Both mechanisms are likely to play key roles in maintaining the parent-of-origin-dependent expression of imprinted genes during gametogenesis. Therefore, it is possible that complete maternal control of reproduction (i.e., apomixis) could be achieved by modifying the epigenetic program that controls imprinting (Bicknell and Koltunow 2004). This assumption, together with the fact that down-regulation of imprinted genes in the FIS complex mimics aspects of apomictic reproduction, such as autonomous embryo and endosperm development, prompted research aimed at linking their expression to apomictic reproduction. For example, the expression profiles of FIS complex genes were proven to be similar in apomictic and sexual lines of Hieracium (Tucker et al. 2003). Moreover, down-regulation of the Arabidopsis FIE ortholog in Hieracium did not result in any autonomous endosperm development, although its activity was necessary for regular seed formation in both apomictic and sexual lines (Rodrigues et al. 2008). Similarly, gene expression studies of the Hieracium homolog of Arabidopsis, MSI1, in germ lineages of apomictic and sexual lines demonstrated that it is not associated with autonomous seed formation in this species (Rodrigues et al. 2010a). In fact, autonomous endosperm development was not induced in sexual species such as rice and maize by genetic transformation of OsFIE1 (Luo et al. 2009) or by down-regulation of ZmFIE1 and ZmFIE2 (Rodrigues et al. 2010b). Overall, the current findings indicate that the repressive function of the FIS complex is not conserved beyond Arabidopsis and that FIS genes are not directly involved in triggering apomixis in sexual species.
In conclusion, factors delivered by paternal sperm cells could provide molecular cues that link egg cell fertilization to the first zygotic divisions. This scenario leads to the hypothesis that paternal–maternal triggering mechanisms could have a role in the repression of embryo development in the absence of fertilization. Moreover, epigenetic mechanisms of inactivation or down-regulation of genes by DNA methylation could represent another level of control by which the egg cell is maintained in a quiescent state in the absence of fertilization.
Concluding remarks
Recent molecular studies aimed at understanding the basis of apomixis have failed to properly elucidate its central mystery because most apomicts are not agriculturally important crops and do not have agriculturally important relatives. Additionally, no apomicts have been sequenced so far, therefore genome annotation information is still not available. If it is true that zygotic embryogenesis (sexuality) and apomeiotic parthenogenesis (apomixis) follow similar pathways during embryo and seed development, it is also true that specific genes have to be activated, modulated, or silenced in the primary steps of plant reproduction to ensure that functioning embryo sacs develop from apomeiotic rather than meiotic cells. Other genes could be specifically or differentially expressed in sexual and apomictic plants during embryo and endosperm development.
The main approaches that have been followed to study the molecular basis of apomixis address the isolation of genes that prime the expression of apomixis in natural apomicts and/or the identification of genes that mime the features of apomictic pathways when they are deregulated in model sexual systems. To take advantage of this fundamental knowledge of apomixis with the aim of transferring it into sexual crops, three main strategies have been adopted to date: (1) the direct introgression of apomixis into crop plants by means of conventional breeding schemes; (2) the genetic transformation of crop plants by transferring exogenous genes that control the expression of apomixis; and (3) the genetic transformation of crop plants by deregulating the endogenous genes that trigger the expression of apomixis.
Although traditional breeding has taken advantage of crop species with close apomictic relatives, transferring apomixis to sexual plants has been unsuccessful to date (Savidan 2000). The introgression of apomixis into crop species from wild relatives failed mainly because natural apomicts are characterized by hybridity and polyploidy, and the loci controlling apomixis usually have a simple inheritance but a complex structure. In fact, these loci are apparently located in very large chromosomal regions, which make them recalcitrant to recombination-based genetic mapping strategies in addition to complicating their physical cloning. In the case of Tripsacum, the current evidence suggests the presence of barriers in the transfer of apomixis to maize (Leblanc et al. 2009). The greatest progress has been made with the introgression of apomixis into pearl millet from Pennisetum, where a single alien chromosome persists in the most advanced apomictic lines (Singh et al. 2010; Zeng et al. 2011). In the sexual model plant Arabidopsis, for each element of apomixis identified by mutation so far, the penetrance was shown to be generally low and the related candidate genes were never found to be associated with any feature of apomixis in natural apomicts. Most importantly, the three main components of apomixis could not be successfully combined into a single model plant to date. Consequently, although findings in Arabidopsis represent the first steps toward the synthesis of an artificial apomixis system, the engineering of asexual reproduction in Arabidopsis has not yet been accomplished.
Because there are a large number of candidates, it is likely that a cross-check between apomictic species should be carried out to assess both analogous and unique genes. In fact, if some of these genes are truly involved in apomixis, their functions should be conserved in other species. For example, Laspina et al. (2008) carried out a bioinformatics comparison among their own sequences and those reported by Albertini et al. (2004, 2005), identifying five genes with identical annotations. Several of the genes that are differentially expressed in both Paspalum spp. and P. pratensis appear to be involved in an ERK signal transduction cascade, with deviations controlled by a Ras ortholog and phospholipase C. Moreover, to further confirm the involvement of APOSTART in apomixis, partial/complete cDNA fragments showing sequence homology with this gene were isolated in P. squamulatum, Cenchrus ciliaris and H. perforatum. In all three species, an APOSTART member was found expressed specifically in parthenogenic individuals at the stage when embryo develops (Marconi et al. 2013). A similar bioinformatics comparison was also performed between the aposporic species P. simplex and H. perforatum, revealing good candidates for apomixis (located on the ACR, apomixis controlling region of Paspalum, and linked to the HAPPY locus of Hypericum) shared by these two model systems (Schallau et al. 2010; Calderini et al. 2012). In particular, this gene, known in Arabidopsis to be involved in the initiation of DNA replication and to be regulated transcriptionally during cell cycle, is currently under investigations in both species (Galla and Pupilli, personal communication).
We are confident that a novel perspective on the old dilemma of apomixis can emerge not only from a rigorous cross-check among apomixis candidate genes cloned in natural apomictic species but also from a high-throughput analysis of sexual mutants in which pivotal genes control the expression of apomictic components, such as apomeiosis (both apospory and diplospory), parthenogenesis, and/or autonomous endosperm development. The link between apomixis and gene-specific silencing mechanisms, including chromatin remodeling factors or trans-acting and heterochromatic small interfering RNAs involved in both transcriptional and post-transcriptional gene regulation, is beginning to become clear. In fact, merging lines of evidence regarding the role of microRNAs in the control of transcription factors, which act on genes directly involved in the development of embryo sacs, embryos, and seeds, have been reported in Arabidopsis.
Our opinion is that the final steps toward understanding the genetic system controlling apomixis are now being taken, and upcoming research programs will be crucial for reaching a turning point that could actually represent the “year zero” from which apomixis begins to be successfully introduced or mimed in sexual species and hence utilized in the most important crop species to support agricultural and food interests worldwide.
References
Akiyama Y, Conner JA, Goel S, Morishige DT, Mullet JE, Hanna WW, Ozias-Akins P (2004) High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol 134:1733–1741
Albertini E, Barcaccia G (2007) Apomeiosis in Poa pratensis L. and Medicago sativa L. In: Horandl E, Grossniklaus U, Sharbel T, van Dijk P (eds) Apomixis: evolution, mechanisms and perspectives. Gantner, Ruggel [Regnum Veg. 147], pp 93–116
Albertini E, Porceddu A, Ferranti F, Reale L, Barcaccia G, Falcinelli M (2001) Apospory and parthenogenesis may be uncoupled in Poa pratensis L: cytological and genetic evidences. Sex Plant Reprod 14:213–217
Albertini E, Marconi G, Barcaccia G, Raggi L, Falcinelli M (2004) Isolation of candidate genes for apomixis in Poa pratensis L. Plant Mol Biol 56:879–894
Albertini E, Marconi G, Reale L, Barcaccia G, Porceddu A, Ferranti F, Falcinelli M (2005) SERK and APOSTART: candidate genes for apomixis in Poa pratensis L. Plant Physiol 138:2185–2199
Albertini E, Barcaccia G, Mazzucato A, Sharbel TF, Falcinelli M (2010) Apomixis in the era of biotechnology. In: Pua EC, Davey MR (eds) Plant developmental biology—biotechnological perspectives, vol 1. Springer, Berlin, pp 405–436
Antoine AF, Faure J-E, Cordeiro C, Dumas C, Rougier M, Feijó JA (2000) A calcium influx is triggered and propagates in the zygote as a warfront during in vitro fertilization of flowering plants. Proc Natl Acad Sci USA 97:10643–10648
Asker SE, Jerling L (1992) Apomixis in plants. CRC Press, Boca Raton, FL
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159
Bajon C, Horlow C, Motamayor J, Sauvanet A, Robert D (1999) Megasporogenesis in Arabidopsis thaliana L: an ultrastructural study. Sex Plant Reprod 12:99–109
Barcaccia G, Mazzucato A, Albertini E, Zethof J, Pezzotti M, Gerats A, Falcinelli M (1998) Inheritance of parthenogenesis in Poa pratensis L: auxin test and AFLP linkage analyses support monogenic control. Theor Appl Genet 97:74–82
Barcaccia G, Mazzucato A, Falcinelli M (2000) Inheritance of apomictic seed production in Kentucky bluegrass (Poa pratensis L). J New Seeds 2:43–58
Barcaccia G, Arzenton F, Sharbel TF, Varotto S, Parrini P, Lucchin M (2006) Genetic diversity and reproductive biology in ecotypes of the facultative apomict Hypericum perforatum L. Heredity 96:322–334
Barcaccia G, Baumlein H, Sharbel TF (2007) Apomixis in St. John’s wort: an overview and glimpse towards the future. In: Hörandl E, Grossniklaus U, Van Dijk P, Sharbel TF (eds) Apomixis. Mechanisms and perspectives. International Association of Plant Taxonomy, Koeltz Scientific Books, Vienna, pp 259–280
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933
Bicknell RA, Koltunow AM (2004) Understanding apomixis: recent advances and remaining conundrums. Plant Cell 16:S228–S245
Bicknell RA, Borst NK, Koltunow AM (2000) Monogenic inheritance of apomixis in two Hieracium species with distinct developmental mechanisms. Heredity 84:228–237
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, Custers JB, van Lookeren Campagne MM (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749
Calderini O, Martinez E, Hojsgaard D, Quarin C, Paolocci F, Caceres ME, Donnison I, Baümlein H, Pupilli F (2012) Comparative genomic analysis of the apomixis locus in Paspalum spp. J Basic Appl Genet 23:61–62
Cao X, Jacobsen SE (2002) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12:1138–1144
Carman JG (1997) Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Linn Soc 61:51–94
Catanach AS, Erasmuson SK, Podivinsky E, Jordan BR, Bicknell R (2006) Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc Natl Acad Sci USA 103:18650–18655
Cervigni GD, Paniego N, Pessino S, Selva JP, Zappacosta D, Spangenberg G, Echenique VC (2008) Gene expression in diplosporous and sexual Eragrostis curvula genotypes with differing ploidy levels. Plant Mol Biol 67:11–23
Chaudhury AM, Peacock JW (1993) Approaches to isolating apomictic mutants in Arabidopsis thaliana: prospects and progress. In: Khush GS (ed) Apomixis: exploiting hybrid vigour in rice. International Rice Research Institute, Manila, pp 66–71
Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94:4223–4228
Chen L, Miyazaki C, Kojima A, Saito A, Adachi T (1999) Isolation and characterization of a gene expressed during early embryo sac development in apomictic Guinea grass (Panicum maximum). J Plant Physiol 154:55–62
Chen L, Guan L, Seo M, Hoffmann F, Adachi T (2005) Developmental expression of ASG-1 during gametogenesis in apomictic guinea grass (Panicum maximum). J Plant Physiol 162:1141–1148
Choi YH, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110:33–42
Citterio S, Varotto S, Albertini E, Feltrin E, Soattin M, Marconi G, Sgorbati S, Lucchin M, Barcaccia G (2005) Alfalfa Mob1-like proteins are expressed in reproductive organs during meiosis and gametogenesis. Plant Mol Biol 58:789–807
Citterio S, Piatti S, Albertini E, Aina R, Varotto S, Barcaccia G (2006) Alfalfa Mob1-like proteins are involved in cell proliferation and are localized in the cell division plane during cytokinesis. Exp Cell Res 312:1050–1064
Conner JA, Goel S, Gunawan G, Cordonnier-Pratt MM, Johnson VE, Liang C, Wang H, Pratt LH, Mullett JE, DeBarry J, Yang L, Bennetzen JL, Klein PE, Ozias-Akins P (2008) Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol 147:1396–1411
Conner J, Gunawan G, Ozias-Akins P (2013) Recombination within the apospory specific genomic region leads to the uncoupling of apomixis components in Cenchrus ciliaris. Planta. doi:10.1007/s00425-013-1873-5
Corral JM, Vogel H, Aliyu OM, Hensel G, Thiel T, Kumlehn J, Sharbel T (2013) APOLLO: a candidate allele linked to apomeiosis in Boechera. In: Proceedings of the international plant and animal genome conferences, San Diego, CA. https://pag.confex.com/pag/xxi/webprogram/Paper5196.html
Curtis MD, Grossniklaus U (2007) Amphimixis and apomixis: two sides of the same coin! In: Hörandl E, Grossniklaus U, Van Dijk P, Sharbel TF (eds) Apomixis: evolution, mechanisms and perspectives. International Association of Plant Taxonomy, Koeltz Scientific Books, Vienna, pp 37–62
Curtis MD, Grossniklaus U (2008) Molecular control of autonomous embryo and endosperm development. Sex Plant Reprod 21:79–88
d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R (2009) Turning meiosis into mitosis. PLoS Biol 7:e1000124
d’Erfurth I, Cromer L, Jolivet S, Girard C, Horlow C, Sun Y, To JPC, Berchowitz LE, Copenhaver GP, Mercier R (2010) The CYCLIN-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet 6:e1000989
Diévart A, Clark SE (2004) LRR-containing receptors regulating plant development and defense. Development 131:251–261
do Valle CB, Savidan YH (1996) Genetics, cytogenetics, and reproductive biology of Brachiaria. In: Miles JW, Maass BL, do Valle CB (eds) Brachiaria: biology, agronomy and improvement. CIAT, Cali, Colombia, pp 147–163
Dwivedi KK, Bhat BV, Gupta MG, Bhat SR, Bhat V (2007) Identification of a SCAR marker linked to apomixis in buffelgrass (Cenchrus ciliaris L.). Plant Sci 172:788–795
Dwivedi SL, Perotti E, Hari D, Upadhyaya HD, Ortiz R (2010) Sexual and apomictic plant reproduction in the genomics era: exploring the mechanisms potentially useful in crop plants. Sex Plant Reprod 23:265–279
Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429:776–780
Ebina M, Nakagawa H, Yamamoto T, Araya H, Tsuruta S-I, Takahara M, Nakajima K (2005) Co-segregation of AFLP and RAPD markers to apospory in Guineagrass (Panicum maximum Jacq.). Grassland Sci 51:71–78
Evans MM (2007) The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19:46–62
Felitti SA, Seijo JG, González AM, Podio M, Laspina NV, Siena L, Ortiz JPA, Pessino SC (2011) Expression of lorelei-like genes in aposporous and sexual Paspalum notatum plants. Plant Mol Biol 77:337–354
Galla G, Barcaccia G (2012) Understanding the molecular basis of apomixis in Hypericum perforatum: de novo sequencing and annotation of the flower organ transcriptome. In: Proceedings of the 2nd global congress of plant reproductive biology, Pécs (Hungary), 15–18 Apr 2012
Galla G, Barcaccia G, Schallau A, Puente Molins M, Bäumlein H, Sharbel TF (2010) The cytohistological basis of apospory in Hypericum perforatum L. Sex Plant Reprod 24:47–61
Galla G, Volpato M, Sharbel TF, Barcaccia G (2013) Computational identification of conserved microRNAs and their putative targets in Hypericum perforatum L. flower transcriptomes. Plant Reprod. doi:10.1007/s00497-013-0227-6
Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D (2010) Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22:3249–3267
Goel S, Chen Z, Akiyama Y, Conner JA, Basu M, Gualtieri G, Hanna WW, Ozias-Akins P (2006) Comparative physical mapping of the apospory-specific genomic region in two apomictic grasses: Pennisetum squamulatum and Cenchrus ciliaris. Genetics 173:389–400
Grimanelli D, Leblanc O, Espinosa E, Perotti E, Gonzalez de Leon D, Savidan Y (1998) Non-mendelian transmission of apomixis in maize-Tripsacum hybrids caused by a transmission ratio distortion. Heredity 80:40–47
Grimanelli D, Leblanc O, Perotti E, Grossniklaus U (2001) Developmental genetics of gametophytic apomixis. Trends Genet 17:597–604
Grimanelli D, García M, Kaszas E, Perotti E, Leblanc O (2003) Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 165:1521–1531
Gross-Hardt R, Lenhard M, Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev 16:1129–1138
Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280:446–450
Grossniklaus U, Spillane C, Page DR, Köhler C (2001a) Genomic imprinting and seed development: endosperm formation with and without sex. Curr Opin Plant Biol 4:21–27
Grossniklaus U, Nogler GA, van Dijk PJ (2001b) The genetic control of gametophytic apomixis. Plant Cell 13:1491–1498
Guerin J, Rossel JB, Robert S, Tsuchiya T, Koltunow A (2000) A DEFICIENS homologue is down-regulated during apomictic initiation in ovules of Hieracium. Planta 210:914–920
Guitton EA, Berger F (2005) Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenic embryos in Arabidopsis. Curr Biol 15:750–754
Hagberg A, Hagberg G (1980) High frequency of spontaneous haploids in the progeny of an induced mutation in barley. Hereditas 93:341–343
Hecht V, Vielle-Calzada J-P, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, de Vries SC (2001) The Arabidopsis somatic embryogenesis receptor kinase 1 (SERK1) gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127:803–816
Henderson JT, Li H-C, Rider SD, Mordhorst AP, Romero-Severson J, Cheng J-C, Robey J, Sung ZR, de Vries SC, Ogas J (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134:995–1005
Hirabayashi S, Nakagawa K, Sumita K, Hidaka S, Kawai T, Ikeda M, Kawata A, Ohno K, Hata Y (2008) Threonine 74 of MOB1 is a putative key phosphorylation site by MST2 to form the scaffold to activate nuclear Dbf2-related kinase 1. Oncogene 27:4281–4292
Hojsgaard DH, Schegg E, Valls JFM, Martinez EJ, Quarin CL (2008) Sexuality, apomixis, ploidy levels, and genomic relationships among four Paspalum species of the subgenus Anachyris (Poaceae). Flora 203:535–547
Hojsgaard DH, Martínez EJ, Quarin CL (2013) Competition between meiotic and apomictic pathways during ovule and seed development results in clonality. New Phytol 197:336–347
Huo H, Conner JA, Ozias-Akins P (2009) Genetic mapping of the apospory-specific genomic region (ASGR) in Pennisetum squamulatum using retrotransposon-based molecular markers. Theor Appl Genet 119:199–212
Ingouff M, Haseloff J, Berger F (2005) Polycomb group genes control developmental timing of endosperm. Plant J 42:663–674
Jessup RW, Burson BL, Burow GB, Wang YWCC, Li Z, Paterson AH, Hussey MA (2002) Disomic inheritance, suppressed recombination, and allelic interactions govern apospory in buffelgrass as revealed by genome mapping. Crop Sci 42:1688–1694
Jessup RW, Burson BL, Burow G, Wang Y-W, Chang C, Li Z, Paterson AH, Hussey MA (2003) Segmental allotetraploidy and allelic interactions in buffelgrass (Pennisetum ciliare (L.) Link syn. Cenchrus ciliaris L.) as revealed by genome mapping. Genome 46:304–313
Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W (2008) A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol 18:1680–1686
Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F (2008) Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol 6:1693–1705
Kaushal P, Malaviya DR, Roy AK, Pathak S, Agrawal A, Khare A, Siddiqui SA (2008) Reproductive pathways of seed development in apomictic guinea grass (Panicum maximum Jacq.) reveal uncoupling of apomixis components. Euphytica 164:81–92
Klattenhoff C, Theurkauf W (2008) Biogenesis and germline functions of piRNAs. Development 135:3–9
Köhler C, Henning L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17:1540–1553
Koltunow AM, Grossniklaus U (2003) Apomixis: a developmental perspective. Annu Rev Plant Biol 54:547–574
Koltunow AM, Bicknell RA, Chaudhury AM (1995) Apomixis: molecular strategies for the generation of genetically identical seeds without fertilization. Plant Physiol 108:1345–1352
Koltunow AMG, Johnson SD, Rodrigues JCM, Okada T, Hu Y, Tsuchiya T, Wilson S, Fletcher P, Ito K, Suzuki G et al (2011) Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella, in which two dominant loci function to enable apomixis. Plant J 66:890–902
Koltunow AMG, Ozias-Akins P, Siddiqi I (2013) Apomixis. In: Becraft PW (ed) Seed genomics. Wiley-Blackwell, Oxford, pp 83–110
Labombarda P, Busti A, Càceres ME, Quarìn CL, Pupilli F, Arcioni S (2002) An AFLP marker tightly linked to apomixis reveals hemizygosity in a portion of the apomixis-controlling locus in Paspalum simplex. Genome 45:513–519
Lacadena J-R (1974) Spontaneous and induced parthenogenesis and androgenesis. In: Kasha KJ (ed) Haploids in higher plants: advances and potential. The University of Guelph, Guelph, pp 13–32
Lacerda ALM, Alencar Dusi DM, Alves ER, Rodrigues JCM, Gomes ACMM, Carneiro VTC (2013) Expression analyses of Brachiaria brizantha genes encoding ribosomal proteins BbrizRPS8, BbrizRPS15a, and BbrizRPL41 during development of ovaries and anthers. Protoplasma 250:505–514
Laspina NV, Vega T, Seijo G, González AM, Martelotto LG, Stein J, Podio M, Ortiz JPA, Echenique VC, Quarin CL, Pessino SC (2008) Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Mol Biol 67:615–628
Leblanc O, Grimanelli D, Gonzalez-de-Leon D, Savidan Y (1995) Detection of the apomictic mode of reproduction in maize-Tripsacum hybrids using maize RFLP markers. Theor Appl Genet 90:1198–1203
Leblanc O, Armstead I, Pessino S, Ortiz JPA, Evans C, do Valle C, Hayward MD (1997) Nonradioactive mRNA fingerprinting to visualize gene expression in mature ovaries of Brachiaria hybrids derived from B. brizantha, an apomictic tropical forage. Plant Sci 126:49–58
Leblanc O, Grimanelli D, Hernandez-Rodriguez M, Galindo PA, Soriano-Martinez AM, Perotti E (2009) Seed development and inheritance studies in apomictic maize-Tripsacum hybrids reveal barriers for the transfer of apomixis into sexual crops. Intl J Dev Biol 53:585–596
Li L-C, Qin G-J, Tsuge T, Hou X-H, Ding M-Y, Aoyama T, Oka A, Chen Z, Gu H, de Zhao Y, Qu L-J (2008) SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 179:751–764
Lieber D, Lora J, Schrempp S, Lenhard M, Laux T (2011) Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr Biol 21:1009–1017
Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205
Lovell JT, Aliyu OM, Mau M, Schranz ME, Koch M, Kiefer C, Song B-H, Mitchell-Olds T, Sharbel TF (2013) On the origin and evolution of apomixis in Boechera. Plant Reprod. doi:10.1007/s00497-013-0218-7
Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96:296–301
Luo M, Platten D, Chaudhury AM, Peacock WJ, Dennis ES (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2:711–723
Marconi G, Sharbel T, Colombo L, Masiero S, Barcaccia G, Galla G, Conner J, Ozias-Akins P, Albertini E (2013) Differential expression in sexual and apomictic genotypes of three model species strengthens a crucial role of APOSTART in the formation of seeds by apomixis. In: International plant and animal genome conferences, San Diego, CA, USA, 11–16 Jan 2013, Abstract W031. https://pag.confex.com/pag/xxi/webprogram/Paper5196.html
Martinez EJ, Hopp HE, Stein J, Ortiz JPA, Quarin CL (2003) Genetic characterization of apospory in tetraploid Paspalum notatum based on the identification of linked molecular markers. Mol Breed 12:319–327
Matzk F (1996) The ‘Salmon System’ of wheat—a suitable model for apomixis research. Hereditas 125:299–304
Matzk F, Meister A, Brutovska R, Schubert I (2001) Reconstruction of reproductive diversity in Hypericum perforatum L. opens novel strategies to manage apomixis. Plant J 26:275–282
Matzk F, Prodanovic S, Baumlein H, Schubert I (2005) The inheritance of apomixis in Poa pratensis confirms a five locus model with differences in gene expressivity and penetrance. Plant Cell 17:13–24
Miyazaki S, Ito M (2006) Calcium signals for egg activation in mammals. J Pharmacol Sci 100:545–552
Muntzing A (1940) Further studies on apoximis and sexuality in Poa. Hereditas 27:115–190
Naumova T (1992) Apomixis in angiosperms: nucellar and integumentary embryony. CRC Press, Boca Raton, FL
Nogler GA (1984) Gametophytic apomixis. In: Johri BM (ed) Embryology of angiosperms. Springer, Berlin, pp 475–518
Nogler GA (1995) Genetics of apomixis in Ranunculus auricomus. VI. Epilogue. Bot Helv 105:111–115
Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15:1728–1739
Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N (2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during the male and female sporogenesis in rice. Plant Cell 19:2583–2594
Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N (2008) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotis mitosis and meiosis during the male and female sporogenesis in rice. Plant Cell 19:2583–2594
Noyes RD, Rieseberg LH (2000) Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 155:379–390
Noyes RD, Baker R, Mai B (2007) Mendelian segregation for two-factor apomixis in Erigeron annuus (Asteraceae). Heredity 98:92–98
Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11:407–416
Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464:628–632
Ozias-Akins P (2006) Apomixis: developmental characteristics and genetics. Crit Rev Plant Sci 25:199–214
Ozias-Akins P, van Dijk PJ (2007) Mendelian genetics of apomixis. Annu Rev Genet 41:509–537
Ozias-Akins P, Roche D, Hanna WW (1998) Tight clustering and hemizygosity of apomxis-linked markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc Natl Acad Sci USA 95:5127–5132
Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V (2009) Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324:1684–1689
Pawlowski WP, Wang C-JR, Golubovskaya IN, Szymaniak JM, Shi L, Hamant O, Zhu T, Harper L, Sheridan WF, Cande WZ (2009) Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc Natl Acad Sci USA 106:3603–3608
Peacock JP (1992) Genetic engineering and mutagenesis for apomixis in rice. Apomixis Newsl 4:3–7
Pessino SC, Ortiz JPA, Leblanc O, do Valle CB, Hayward MD (1997) Identification of a maize linkage group related to apomixis in Brachiaria. Theor Appl Genet 94:439–444
Pessino SC, Evans C, Ortiz JPA, Armstead I, do Valle CB, Hayward MD (1998) A genetic map of the apospory-region in Brachiaria hybrids: identification of two markers closely associated with the trait. Hereditas 128:153–158
Pessino SC, Espinoza F, Martinez EJ, Ortiz JPA, Valle EM, Quarin CL (2001) Isolation of cDNA clones differentially expressed in flowers of apomictic and sexual Paspalum notatum. Hereditas 134:35–42
Pillot M, Autran D, Leblanc O, Grimanelli D (2010a) A role for CHROMOMETHYLASE3 in mediating transposon and euchromatin silencing during egg cell reprogramming in Arabidopsis. Plant Signal Behav 5:1167–1170
Pillot M, Baroux C, Vazquez MA, Autran D, Leblanc O, Vielle-Calzada JP, Grossniklaus U, Grimanelli D (2010b) Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22:307–320
Podio M, Rodriguez MP, Felitti S, Stein J, Martinez E, Siena LA, Quarin CL, Pessino SC, Ortiz JPA (2012) Sequence characterization, in silico mapping and cytosine methylation analysis of markers linked to apospory in Paspalum notatum. Genet Mol Biol 35:827–837
Polegri L, Calderini O, Arcioni S, Pupilli F (2010) Specific expression of apomixis-linked alleles revealed by comparative transcriptomic analysis of sexual and apomictic Paspalum simplex Morong flowers. J Exp Bot 61:1869–1883
Pupilli F, Barcaccia G (2012) Cloning plants by seeds: inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. J Biotechnol 159:291–311
Pupilli F, Labombarda P, Caceres ME, Quarin CL, Arcioni S (2001) The chromosome segment related to apomixis in Paspalum simplex is homoeologous to the telomeric region of the long arm of rice chromosome 12. Mol Breed 8:53–61
Pupilli F, Martinez EJ, Busti A, Calderini O, Quarìn CL, Arcioni S (2004) Comparative mapping reveals partial conservation of synteny at the apomixis locus in Paspalum spp. Mol Genet Genomics 270:539–548
Qiu Y, Liu R, Xie C, Russell S, Tian H (2008) Calcium changes during megasporogenesis and megaspore degeneration in lettuce (Lactuca sativa L). Sex Plant Reprod 21:197–204
Ravi M, Chan SWL (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464:615–618
Ravi M, Marimuthu MP, Siddiqi I (2008) Gamete formation without meiosis in Arabidopsis. Nature 451:1121–1124
Rhoades MM, Dempsey E (1966) Induction of chromosome doubling at meiosis by the elongate gene in maize. Genetics 54:505–522
Roche D, Cong P, Chen Z, Hanna WW, Gustine DL, Sherwood RT, Ozias-Akins P (1999) An apospory-specific genomic region is conserved between Buffelgrass (Cenchrus ciliaris L.) and Pennisetum squamulatum Fresen. Plant J 19:203–208
Roche D, Chen Z, Hanna WW, Ozias-Akins P (2001) Non-Mendelian transmission of an apospory-specific genomic region in a reciprocal cross between sexual pearl millet (Pennisetum glaucum) and an apomictic F1 (P. glaucum × P. squamulatum). Sex Plant Reprod 13:217–223
Rodrigues JCM, Cabral GB, Dusi DMA, de Mello LV, Rigden DJ, Carneiro VTC (2003) Identification of differentially expressed cDNA sequences in ovaries of sexual and apomictic plants of Brachiaria brizantha. Plant Mol Biol 53:745–757
Rodrigues JCM, Tucker MR, Johnson SD, Hrmova M, Koltunow AMG (2008) Sexual and apomictic seed formation in Hieracium requires the plant polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM. Plant Cell 20:2372–2386
Rodrigues JCM, Luo M, Berger F, Koltunow AMG (2010a) Polycomb group gene function in sexual and asexual seed development in angiosperms. Sex Plant Reprod 23:123–133
Rodrigues JCM, Okada T, Johnson SD, Koltunow AM (2010b) A multicopy suppressor of IRA1 (MSI1) homologue is not associated with the switch to autonomous seed development in apomictic (asexual) Hieracium plants. Plant Sci 179:590–597
Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24:1010–1021
Savidan Y (2000) Apomixis: genetics and breeding. Plant Breed Rev 18:13–86
Schallau A, Arzenton F, Johnston AJ, Hähnel U, Koszegi D, Blattner FR, Altschmied L, Haberer G, Barcaccia G, Bäumlein H (2010) Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J 62:773–784
Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96:11664–11669
Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U (2011) Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol 9:e1001155
Schranz ME, Kantama L, de Jong H, Mitchell-Olds T (2006) Asexual reproduction in a close relative of Arabidopsis: a genetic investigation of apomixis in Boechera (Brassicaceae). New Phytol 171:425–438
Schwartz YB, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8:9–22
Selva JP, Pessino S, Meier M, Echenique V (2012) Identification of candidate genes related to polyploidy and/or apomixis in Eragrostis curvula. Am J Plant Sci 3:403–416
Sharbel TF, Voigt ML, Corral JM, Thiel T, Varshney A, Kumlehn J, Vogel H, Rotter B (2009) Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J 58:870–882
Sharbel TF, Voigt ML, Corral JM, Galla G, Kumlehn J, Klukas C, Schreiber F, Vogel H, Rotter B (2010) Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 22:655–671
Sheridan WF, Avalkina NA, Shamrov II, Batygina TB, Golubovskaya IN (1996) The mac1 gene: controlling the commitment to the meiotic pathway in maize. Genetics 142:1009–1020
Sheridan WF, Golubeva EA, Abrhamova LI, Golubovskaya IN (1999) The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics 153:933–941
Sherwood RT, Berg CC, Young BA (1994) Inheritance of apospory in buffelgrass. Crop Sci 34:1490–1494
Sieber P, Gheyselinck J, Gross-Hardt R, Laux T, Grossniklaus U, Schneitz K (2004) Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol 273:321–334
Silveira ÉD, Guimarães LA, Alencar Dusi DM, Silva FR, Martins NF, Carmo Costa MM, Alves-Ferreira M, Carneiro VTC (2012) Expressed sequence-tag analysis of ovaries of Brachiaria brizantha reveals genes associated with the early steps of embryo sac differentiation of apomictic plants. Plant Cell Rep 31:403–416
Silvertown J (2008) The evolutionary maintenance of sexual reproduction: evidence from the ecological distribution of asexual reproduction in clonal plants. Intl J Plant Sci 169:157–168
Singh M, Burson BL, Finlayson SA (2007) Isolation of candidate genes for apomictic development in buffelgrass (Pennisetum ciliare). Plant Mol Biol 64:673–682
Singh M, Conner JA, Zeng Y-J, Hanna WW, Johnson VE, Ozias-Akins P (2010) Characterization of apomictic BC7 and BC8 pearl millet: meiotic chromosome behavior and construction of an ASGR-carrier chromosome-specific library. Crop Sci 50:892–902
Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D (2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23:443–458
Slotkin RK, Vaughn M, Filipe Borges F, Tanurdžić M, Becker JD, Feijó JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–472
Spillane C, Curtis MD, Grossniklaus U (2004) Apomixis technology development—virgin births in farmers’ fields? Nat Biotechnol 22:687–691
Stein J, Pessino SC, Martínez EJ, Rodriguez MP, Siena LA, Quarìn CL, Ortiz JPA (2007) A genetic map of tetraploid Paspalum notatum Flügge (bahiagrass) based on single-dose molecular markers. Mol Breed 20:153–166
Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98:11806–11811
Sundaresan V, Alandete-Saez M (2010) Pattern formation in miniature: the female gametophyte of flowering plants. Development 137:179–189
Tas ICQ, van Dijk PJ (1999) Crosses between sexual and apomictic dandelions (Taraxacum). I. The inheritance of apomixis. Heredity 83:707–714
Tucker MR, Koltunow AM (2009) Sexual and asexual (apomictic) seed development in flowering plants: molecular, morphological and evolutionary relationships. Funct Plant Biol 36:490–504
Tucker MR, Araujo AC, Paech NA, Hecht V, Schmidt ED, Rossell JB, De Vries SC, Koltunow AM (2003) Sexual and apomictic reproduction in Hieracium subgenus pilosella are closely interrelated developmental pathways. Plant Cell 15:1524–1537
Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM (2012) Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139:1399–1404
van Dijk PJ, Bakx-Schotman JMT (2004) Formation of unreduced megaspores (diplospory) in apomictic dandelions (Taraxacum officinale, s.l.) is controlled by a sex-specific dominant locus. Genetics 166:483–492
van Dijk PJ, Tas ICQ, Falque M, Bakx-Schotman T (1999) Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 83:715–721
van Dijk PJ, van Baarlen P, de Jong H (2003) The occurrence of phenotypically complementary apomixis-recombinants in crosses between sexual and apomictic dandelions (Taraxacum officinale). Sex Plant Reprod 16:71–76
Van Ex F, Jacob Y, Martienssen RA (2011) Multiple roles for small RNAs during plant reproduction. Curr Opin Plant Biol 14:588–593
Vaucheret H (2008) Plant argonautes. Trends Plant Sci 13:350–358
Vielle-Calzada JP, Crane CF, Stelly DM (1996) Apomixis: the asexual revolution. Science 274:1322–1323
Vijverberg K, van Dijk PJ (2007) Genetic linkage mapping of apomixis loci. In: Hörandl E, Grossniklaus U, van Dijk P, Sharbel TF (eds) Apomixis. Evolution, mechanisms and perspectives. International Association of Plant Taxonomy, Koeltz Scientific Books, Vienna, pp 137–158
Vijverberg K, Van der Hulst RGM, Lindhout P, van Dijk PJ (2004) A genetic linkage map of the diplosporous chromosomal region in Taraxacum officinale (common dandelion; Asteraceae). Theor Appl Genet 108:725–732
Vijverberg K, Milanovic S, Bakx-Schotman T, van Dijk PJ (2010) Genetic fine mapping of DIPLOSPOROUS in Taraxucum (dandelion; Asteraceae) indicates a duplicated DIP-gene. BMC Plant Biol 10:154
Vitulo N, Vezzi A, Galla G, Citterio S, Marino G, Ruperti B, Zermiani M, Albertini E, Valle G, Barcaccia G (2007) Characterization and evolution of the cell cycle-associated Mob domain-containing proteins in eukaryotes. Evol Bioinform 3:121–158
Vyskot B, Hobza R (2004) Gender in plants: sex chromosomes are emerging from the fog. Trends Genet 20:432–438
Yamada-Akiyama H, Akiyama Y, Ebina M, Xu Q, Tsuruta S, Yazaki J, Kishimoto N, Kikuchi S, Takahara M, Takamizo T, Sugita S, Nakagawa H (2009) Analysis of expressed sequence tags in apomictic guineagrass (Panicum maximum). J Plant Physiol 166:750–761
Yang W, Sundaresan V (2000) Genetics of gametophyte biogenesis in Arabidopsis. Curr Opin Plant Biol 3:53–57
Zeng Y, Conner J, Ozias-Akins P (2011) Identification of ovule transcripts from the Apospory-Specific Genomic Region (ASGR)-carrier chromosome. BMC Genomics 12:206
Zhao DZ, Wang GF, Speal B, Ma H (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16:2021–2031
Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Herve P, Xue Q, Bennett J (2008) OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J 54:375–387
Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30:349–359
Acknowledgments
Authors want to thank COST Action FA0903 “Harnessing Plant Reproduction for Crop Improvement.” Gianni Barcaccia was supported by the following grant: Research Project of the University of Padova (year 2012), “Transcriptomics of reproductive organs in model species for comparative analysis of the genetic-molecular factors characterizing sexual and apomictic processes” (code: CPDA128282/12).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Dresselhaus.
A contribution to the Special Issue “HAPRECI—Plant Reproduction Research in Europe.”
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Barcaccia, G., Albertini, E. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod 26, 159–179 (2013). https://doi.org/10.1007/s00497-013-0222-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-013-0222-y