Abstract
Background
Ethylene production and signalling play an important role in somatic embryogenesis, especially for species that are recalcitrant in in vitro culture. The AP2/ERF superfamily has been identified and classified in Hevea brasiliensis. This superfamily includes the ERFs involved in response to ethylene. The relative transcript abundance of ethylene biosynthesis genes and of AP2/ERF genes was analysed during somatic embryogenesis for callus lines with different regeneration potential, in order to identify genes regulated during that process.
Results
The analysis of relative transcript abundance was carried out by real-time RT-PCR for 142 genes. The transcripts of ERFs from group I, VII and VIII were abundant at all stages of the somatic embryogenesis process. Forty genetic expression markers for callus regeneration capacity were identified. Fourteen markers were found for proliferating calli and 35 markers for calli at the end of the embryogenesis induction phase. Sixteen markers discriminated between normal and abnormal embryos and, lastly, there were 36 markers of conversion into plantlets. A phylogenetic analysis comparing the sequences of the AP2 domains of Hevea and Arabidopsis genes enabled us to predict the function of 13 expression marker genes.
Conclusions
This first characterization of the AP2/ERF superfamily in Hevea revealed dramatic regulation of the expression of AP2/ERF genes during the somatic embryogenesis process. The gene expression markers of proliferating callus capacity to regenerate plants by somatic embryogenesis should make it possible to predict callus lines suitable to be used for multiplication. Further functional characterization of these markers opens up prospects for discovering specific AP2/ERF functions in the Hevea species for which somatic embryogenesis is difficult.
Similar content being viewed by others
Background
Understanding the molecular mechanisms controlling somatic embryogenesis is crucial, be it biologically or for applications. Indeed, propagating cultivated plants by somatic embryogenesis still remains limited by the efficiency of the procedures in some recalcitrant species or genotypes. Genetic predeterminism of recalcitrance in some woody and herbaceous perennial species has been linked to their response to wounding and their phenolic compound content [1]. The production of ethylene, carbon dioxide and free radicals lies behind the induction of cell defence mechanisms, leading to the oxidation of phenol compounds, the breakdown of cell walls and membrane peroxidation [2]. Adding growth regulators, and the stress induced by in vitro culture, play a leading role in such mechanisms [3]. These phenomena result in the differentiation of undifferentiated cells and in tissue browning, which lead to a loss in embryogenic capacity [2, 4].
The negative effect of ethylene on somatic embryogenesis has been known for a long time [5]. Some super-embryogenic explant cultures of Medicago truncatula revealed the repression of numerous genes, including those involved in ethylene biosynthesis and signalling [6]. In alfalfa, loss of embryogenic capacity following thidiazuron application is linked to the induction of an ethylene biosynthesis gene [7]. Ethylene also induces some stress factors conducive to the acquisition of somatic embryogenesis capacities [8, 9], and embryo maturation [10]. Abscisic acid and methyl jasmonate are regulators of ethylene biosynthesis during somatic embryogenesis in Medicago sativa[11]. Some members of the ETHYLENE RESPONSE FACTOR (ERF) family are involved in response to in vitro stress and in the regulation of developmental processes. In Medicago truncatula, the SOMATIC EMBRYO RELATED FACTOR 1 gene (MtSERF1) is induced by ethylene and may act under the influence of WUSCHEL (WUS), whose fixation sites are found on the SERF1 gene promoter [12]. ENHANCER OF SHOOT REGENERATION1 (ESR1) of Arabidopsis is induced by cytokinins to regulate the start of shoot regeneration [13]. DORNROSCHEN (DRN)/ESR1 plays a role in meristem and organ development and consequently in shoot regeneration [14]. ESR2 expression confers “cytokinin-independent shoot regeneration” through transcriptional regulation of the CUP-SHAPED COTYLEDON 1 gene (CUC1) [15]. When interacting with other transcription factors, the induction of CALLUS EXPRESSING FACTOR1 (CEF1) through stress would appear to disrupt auxin/cytokinin homeostasis in Nicotiana tabacum. Of the numerous other transcription factors involved in embryogenesis and organ development ([16] for a review), several members of the APETALA2 family (AP2) play a major role [17]. Like ERFs, this family belongs to the AP2/ERF superfamily. For example, BABY BOOM (BBM) is known for its role in cell proliferation and morphogenesis during embryogenesis [18]. AINTEGUMENTA (ANT) is involved in ovule development and in the growth of floral organs [19]. WRINKLED (WRl1) is involved in regulating storage metabolism in seeds [20, 21]. The ANT subfamily includes several members called ANT-like genes or AILs. The initiation of post-embryonic shoot organs takes place in the shoot apical meristem, also involving several members of the AP2 family (ANT, AIL6/PLT3 and AIL7/PLT7) [22].
Hevea brasiliensis is a particularly difficult species which has led to numerous micropropagation studies [23]. This cross-fertilizing species is cloned by budding in the absence of any other efficient vegetative propagation techniques for own-rooted plants. Two somatic embryogenesis methods have been developed for Hevea ([23–25] for a review). The first is somatic embryogenesis on primary callus (SEP) obtained from fragments of the internal integument of immature seeds or anthers [26–28]. This method is effective for around twenty Hevea clones following numerous culture medium and atmosphere studies [25, 29]. However, primary calli are subject to browning due to high ethylene and carbon dioxide release, which leads to a low callus multiplication rate [30]. In vitro plantlets produced by SEP are usually of good quality with better growth and latex production than budded clones [31]. A second method was developed from friable callus maintained over the long term with a view to large-scale multiplication. This method has evolved with the use of fragments of embryos derived from SEP in order to rapidly establish embryogenic friable callus lines [29, 32]. Lastly, cryopreservation of friable calli has been incorporated into the process to limit tissue proliferation and, thereby, the risks linked to somaclonal variation [33]. However, this indirect secondary somatic embryogenesis process is restricted to just a few clones and the embryo and plant regeneration capacity is variable. The absence of plant regeneration potential in some friable callus lines has been linked to early vacuolization of cells in the embryogenic globules [34]. Some transcriptional changes have also been reported for certain Hevea callus lines with different embryogenic potentials [34].
A study on the role of ethylene in stimulating latex production led to the characterization of ethylene biosynthesis and signalling genes and more recently to the identification of the different members of the AP2/ERF superfamily [35–38]. The AP2/ERF superfamily contains 173 members, of which 142 are classed in families and groups [35]. The AP2 family contains 20 genes organized in two subfamilies (8 ANT and 12 AP2 genes). The ERF family is divided into 10 groups comprising a total of 115 genes. Lastly, there remains the RAV family with four genes and the soloists with 3 genes. Based on this knowledge, our study set out to gain a clearer understanding of the regulation of ethylene biosynthesis and signalling genes, including ERFs, along with the members of the AP2 and RAV families during the somatic embryogenesis process in Hevea. The relative transcript abundance analysis for these genes was carried out by real-time RT-PCR on friable callus lines with different embryogenic potential [33], on normal and abnormal somatic embryos, and on different young plant tissues. The gene expression profiles showed that several genes were markers of embryogenic potential for the lines in proliferating calli and in calli induced by embryogenesis, and also markers of somatic embryo quality. Prediction of the function of those marker genes provides a dynamic understanding of the somatic embryogenesis process.
Results
Morphogenetic potential of callus lines with different somatic embryogenesis capacities
The secondary indirect somatic embryogenesis process is described in Figure 1 (cf. Materials and Methods). Primary somatic embryogenesis is induced from inner integument of immature seed as maternal tissue (Figure 2A). Compact callus-bearing mature somatic embryos were obtained after three subcultures (Figure 2B). Friable callus lines were established from fragments of somatic embryo. The proliferating friable calli grown on the ENT medium were fairly comparable whatever their embryogenic capacity (Figure 2D). After four weeks’ culture on EXP, the calli of the non-embryogenic (Figure 2J) and embryogenic (Figure 2H) lines were more hydric and whitish than those of the regenerant line (Figure 2E). Callus growth was slowed during embryo regeneration in the RITA culture system on DEV medium. The calli turned brown in all the lines (Figure 2F, I and K). Browning occurred to the benefit of embryo formation for embryogenic lines (Figure 2F and I). The normal embryos had an embryonic body and two well-developed cotyledons (Figure 2L). The abnormal types of embryos were most numerous: for example, some embryos with a single cotyledon (Figure 2M), or with malformed cotyledons (Figure 2N), and a double embryonic body (Figure 2O) were found. Plantlets derived from normal embryos developed a taproot and a lateral root system, and a stem with leaves within a month (Figure 2G).
Morphology of callus with different embryogenic capacities and somatic embryos. (A) Inner integument of immature seed (scale bar = 5 mm). (B) Compact callus on somatic embryo development medium (MH3) from primary somatic embryogenesis (bar = 5 mm). (C) Embryogenic callus bearing pro-embryo structures (white arrow) on somatic embryo development medium (DEV) (bar = 5 mm). (D) Regenerant line callus on embryogenesis induction medium (ENT) (bar = 1 mm). Calli were from three types of lines on somatic embryogenesis expression medium (EXP): (E) regenerant line callus, (H) embryogenic and non-regenerant line and (J) non-embryogenic line on somatic embryogenesis expression medium (EXP) (bar = 5 mm). Friable callus on embryo development medium (DEV): (F) from regenerant line callus, (I) embryogenic and non-regenerant line and (K) non-embryogenic line (bar = 10 mm). Various types of somatic embryo: (L) normal cotyledonated embryo, and (M, N, O) abnormal somatic embryos (bar = 1 mm). (G) Plantlet from a normal somatic embryo (bar = 10 mm).
The morphogenetic capacities of three friable callus lines were tested up to somatic embryo conversion into plantlets (Table 1). The non-embryogenic line CI04115 (NE) did not produce any somatic embryos. Line CI04106, called embryogenic (E), produced 19.5 somatic embryos per gram of callus, most of which were abnormal (93.54%). Only the regenerant line CI07060 (R) produced a large number of total somatic embryos (590 per gram of callus). The proportion of abnormal embryos was very high for the embryogenic and regenerant lines, at 93.54% and 82.71% respectively. Out of 102 normal embryos derived from the regenerant line transferred to germination medium, 53 developed into plantlets.
Identification of 14 marker genes of somatic embryogenesis capacities during callus proliferation
The relative transcript abundance for ethylene biosynthesis and signalling genes, and for the AP2 and RAV families, was analysed in proliferating calli on ENT medium (Figure 3). Some high levels were found for several genes in each family of genes, except for the RAVs (Figure 3A). The ERFs from groups I, VII and VIII amounted to 8, 15 and 10 genes respectively with levels over 1 in relative value compared to the transcript abundance of the internal control RH2b. The comparison between lines with different embryogenic potential revealed that 14 genes were differentially expressed (Figure 3B). The relative transcript abundance was lower in the calli of the R line than in the E and NE lines for 11 genes, of which three were involved in ethylene biosynthesis and perception (HbSAMS, HbACS2, HbETR1), four were from the ERF family (HbERF-Ia1, HbERF-VI2, HbERF-VIIa4, HbERF-VIIIa3), and 4 were from the AP2 family (HbAP2-1, HbAP2-5, HbAP2-12, HbAP2-18). The transcripts of the HbAP2-11 gene were greatly accumulated in the embryogenic line. Lastly, the transcripts of the HbERF-VIIa23 gene were less abundant in the non-embryogenic line, while, conversely, those of the HbRAV-4 gene were accumulated more in the calli of that line.
Expression profile of 142 genes involved in the ethylene biosynthesis and signalling pathways for callus with different embryogenic capacities grown for 2 weeks on embryogenesis induction medium (ENT). Calli were from three types of lines: (NE) non-embryogenic line (CI04115), (E) embryogenic and non-regenerant line (CI04106), and (R) regenerant line (CI07060). The relative transcript abundances were measured by real-time RT-PCR. Values are the means of the relative transcript abundances of three biological replicates. (Figure 3A) Heat map representation of the expression profile was used for values ranging as follows ≥ 1, 10-1, 10-2, 10-3 and ≤ 10−4 from dark to light green. (Figure 3B) Values of relative transcript abundance in callus of the various lines were analysed with XLSTAT software after log transformation. The statistical analysis was performed with an ANOVA followed by the Student Newman–Keuls test. Values with significantly high relative transcript abundances shown in red and significantly low relative transcript abundances shown in green. The non-significant genes are shown in yellow.
Identification of 35 marker genes of somatic embryogenesis capacities during somatic embryogenesis expression/induction
The calli transferred to a somatic embryogenesis induction medium also strongly expressed a large number of the genes studied for virtually all the families and groups, except for the ERF-V group (Figure 4A). As previously, most of the members of groups I (9 genes), VII (12 genes) and VIII (10 genes) of the ERF family had a high transcript level. The differential expression between calli of the different lines was significant for 35 genes (Figure 4B). Of those genes, 15 had higher transcript accumulation in the regenerant line than in the NE and E lines: 1 ethylene biosynthesis gene (HbSAMS), 11 genes of the ERF family (HbERF-IIIb2, HbERF-IIIc1, HbERF-IIIe3, HbERF-IVa1,HbERF-IVa2, HbERF-IVa3, HbERF-Vb2, HbERF-VI-L1, HbERF-VIIIa7, HbERF-IXb2, HbERF-IXc4), 2 genes of the AP2 family (HbAP2-3, HbAP2-7) and 1 gene of the RAV family (HbRAV-3). Transcripts of the HbERF-VIIIa3 gene were accumulated in the calli of the embryogenic line compared to the other two lines. Lastly, 19 genes comprising 14 ERFs (HbERF-IIIa1, HbERF-Vb3, HbERF-VI3, HbERF-VI4, HbERF-VI-L6, HbERF-VIIa3, HbERF-VIIa4, HbERF-VIIa13, HbERF-VIIa17, HbERF-VIIIa4, HbERF-VIIIa13, HbERF-IXa2, HbERF-IXa3, HbERF-IXa4) and 5 AP2s (HbAP2-4, HbAP2-6, HbAP2-8, HbAP2-11, HbAP2-19) had a lower transcript level in the non-embryogenic line than in the others.
Expression profile of 142 genes involved in the ethylene biosynthesis and signalling pathways for callus with different embryogenic capacities grown for 4 weeks on somatic embryogenesis expression medium (EXP). Calli were from three types of lines: (NE) non-embryogenic line (CI04115), (E) embryogenic and non-regenerant line (CI04106), and (R) regenerant line (CI07060). The relative transcript abundances were measured by real-time RT-PCR. Values are the means of the relative transcript abundance of three biological replicates. (Figure 4A) Heat map representation of the expression profile was used for values ranging as follows ≥ 1, 10-1, 10-2, 10-3 and ≤ 10−4 from dark to light green. (Figure 4B) Values of relative transcript abundance in callus of the various lines were analysed with XLSTAT software after log transformation. The statistical analysis was performed with an ANOVA followed by the Student Newman–Keuls test. Values with significantly high relative transcript abundances shown in red and significantly low relative transcript abundances shown in green. The non-significant genes are shown in yellow.
Sixteen genes were differentially expressed in normal and abnormal somatic embryos
The normal and abnormal somatic embryos produced by the regenerant line revealed a differential expression profile solely for certain members of the ERF family (Figure 5). The gene transcripts belonging to each of the families and groups were highly accumulated, except for the RAV family (Figure 5A). Most of the members of the ERF-I, ERF-VII and ERF-VIII groups were highly expressed in the two types of embryos. The transcript abundance was lower in the normal embryos than in the abnormal embryos for 12 genes belonging to the ERF family (HbERF-Ia1, HbERF-Ib1, HbERF-Ib3, HbERF-Ib6, HbERF-Ib7, HbERF-Ib8, HbERF-IIb5, HbERF-IVa2, HbERF-VIIIa11, HbERF-IXb4, HbERF-IXb8, HbERF-Xb1) (Figure 5B). Conversely, the transcripts of 4 genes were accumulated in the normal embryos (HbERF-VI1, HbERF-VIIa1, HbERF-VIIa3, HbERF-VIIa4).
Expression profile of 142 genes involved in the ethylene biosynthesis and signalling pathways for normal and abnormal embryos from regenerant line (CI07060) callus on embryo development medium (DEV). The relative transcript abundances were measured by real-time RT-PCR. Values are the means of the relative transcript abundance of three biological replicates. (Figure 5A) Heat map representation of the expression profile was used for values ranging as follows ≥ 1, 10-1, 10-2, 10-3and ≤ 10−4 from dark to light green. (Figure 5B) Values of relative transcript abundance in normal and abnormal embryos were analysed with XLSTAT software after log transformation. The statistical analysis was performed with an ANOVA followed by the Student Newman–Keuls test. Values with significantly high relative transcript abundances shown in red and significantly low relative transcript abundances shown in green. The non-significant genes are shown in yellow.
Evolution of transcript abundance during the process of somatic embryogenesis and conversion into plantlets
An overall analysis of the somatic embryogenesis process for the regenerant line using proliferating callus up to the conversion of normal embryos into plantlets showed that the expression of numerous genes was highly modulated (Figure 6). The transcripts of numerous ethylene biosynthesis and transduction genes, and some ERFs from groups I, VII and VIII and of the AP2 family, were highly accumulated (Figure 6A). Thirty-six genes from all the families or groups revealed differential accumulation during the embryogenesis process, except for the ERFs from group X (Figure 6B). Several gene expression profiles were observed. Eleven markers were activated right from the somatic embryogenesis induction phase (HbSAMS, HbACO1, HbEIN3, HbERF-IIIe2, HbERF-VIIa9, HbERF-VIIa23, HbERF-VIIIa3, HbERF-IXb2, HbAP2-5, HbAP2-11, HbAP2-13). The transcripts of eight genes (HbERF-Ib5, HbERF-IIa3, HbERF-IIIa1, HbERF-VIIa1, HbERF-VIIIa14, HbERF-IXa3, HbAP2-1, HbAP2-3) were very abundant in embryogenic tissues, whereas they were significantly less abundant in the tissues of the plantlet. A transient accumulation was seen for two genes (HbERF-Vb2, HbERF-Vb3) in calli induced in embryogenesis on EXP medium, for six genes (HbERF-VIIa3, HbERF-VIIa4, HbERF-VIIa17, HbERF-VIIIa3, HbAP2-2, HbAP2-6) in somatic embryos and for two genes (HbERF-IVa1, HbERF-VIIa18) in both calli on EXP medium and in embryos. The transcripts of three genes (HbERF-IXb2, HbERF-IXb6, HbERF-IXb8) were preferentially accumulated in leaves and, in the case of HbERF-IXc3, in both somatic embryos and leaves. Lastly, the relative transcript abundance was high in all tissues except in embryos, where it decreased significantly for the HbAP2-16 gene, and except in calli, embryos and roots for HbRAV4.
Expression profile of 142 genes involved in the ethylene biosynthesis and signalling pathways for regenerant line (CI07060) callus on embryogenesis induction medium (ENT) and somatic embryogenesis expression medium (EXP), embryos and 3 plantlet organs: (L) Leaf, (S) Stem, (R) Root. The relative transcript abundances were measured by real-time RT-PCR. Values are the means of the relative transcript abundance of three biological replicates. (Figure 6A) Heat map representation of the expression profile was used for values ranging as follows ≥ 1, 10-1, 10-2, 10-3 and ≤ 10−4 from dark to light green. (Figure 6B) Values of relative transcript abundance were analysed with XLSTAT software after log transformation. The statistical analysis was performed with an ANOVA followed by the Student Newman–Keuls test. Values with significantly high relative transcript abundances shown in red and significantly low relative transcript abundances shown in green. The non-significant genes are shown in yellow.
Identification of putative functions for somatic embryogenesis marker genes
Of the fifty-six genes differentially expressed during the somatic embryogenesis process, forty were regulated in the same way when the regenerant line was compared with the other two non-embryogenic or embryogenic lines, and when the normal embryos were compared with the abnormal embryos (Table 2). The ratios between the relative transcript abundance of the regenerant line and the other two lines showed that the markers tended to be under-expressed in the proliferating calli of the regenerant line (from 0.01 to 0.4) and over-expressed in the calli induced in embryogenesis on EXP medium (from 2 to 43 times). Of the sixteen embryo expression markers, twelve were under-expressed and four were over-expressed.
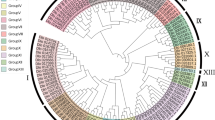
The role of these gene expression markers was first predicted by a reciprocal best hit analysis (BLASTX) of the transcript sequences (Table 2). Roles were mostly found in the response to biotic and abiotic stress, and more precisely in tolerance of dehydration, salinity and cold. The genes of the AP2 and RAV families, along with a small proportion of ERFs, played a role in developmental processes (embryogenesis and flower development). In order to find Hevea potential orthologs to Arabidopsis genes functionally described in the literature, the deduced amino acid sequences of the AP2 domain were analysed for the AP2 family and the different ERF groups (I, III, IV, VI, X) making up the marker genes in Hevea (Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5 and Additional file 6: Figure S6). The summary of the phylogenetic tree analyses can be found in Table 2. Thirteen of the Hevea marker genes proved to be potential orthologs of sixteen Arabidopsis genes including some co-orthologs: HbERF-Ia1 with ERF53, HbERF-IIIc1 with AtCBF/DREB (CBF3/DREB1A, CBF2/DREB1C, CBF1/DREB1B, CBF4/DREB1D, DDF1, DDF2), HbERF-IIIe3 with TINY, HbERF-IVa1 and HbERF-IVa2 with DREB2A, HbERF-VI1 and HbERF-VI2 with CRF2/TMO3, HbERF-Xb1 with RRTF1, HbAP2-1 with BBM, HbAP2-3 with two AINTEGUMENTA-Like genes (AIL7/PLT7 and AIL6/PLT3), HbAP2-7 with AINTEGUMENTA, and HbAP2-12 and HbAP18 with APETALA2.
Discussion
Factors contributing to the loss of embryogenic capacity
Although callus browning occurs late during embryo development in the normal process, early browning in proliferating calli contributes to a loss of their embryogenic capacity by promoting the differentiation of active cells (meristematic and embryogenic) [25]. In Hevea, such browning has been linked to a strong accumulation of oxidized polyphenols in cells and to ethylene production [4, 30, 71]. This study corroborates those findings with the activation of ethylene biosynthesis and signalling genes in proliferating calli of low embryogenic or non-embryogenic lines (Figure 3), whereas this only occurred after induction in embryogenesis on EXP medium for the regenerant line. The gene expression markers of plant regeneration capacity found in proliferating calli belonged to groups VI, VII and VIII of the ERFs (HbERF-VI2, HbERF-VIIa4, HbERF-VIIIa3), which are factors of response to hormonal signals (ethylene, jasmonate, etc.). Other factors generally linked to development were also activated in proliferating calli from the embryogenic or non-embryogenic lines. Four genes from the AP2 family can be noted: HbAP2-1, HbAP2-5, HbAP2-12, HbAP2-18. The phylogenetic analysis between the Hevea and Arabidopsis families led to the prediction of several orthologs. HbAP2-1 and HbAP2-12/HbAP2-18 were found to be potential orthologs of Arabidopsis BBM and AP2 genes, respectively [18, 60]. BBM is preferentially expressed in developing embryos and seeds [18]. Its ectopic expression in Arabidopsis, Brassica and Nicotiana has led to the spontaneous formation of somatic embryos and cotyledon-like structures in seedlings [18, 72]. However, such ectopic expression gives rise to pleiotropic phenotypes such as neoplastic growth, regeneration of plants on a hormone-free medium, and an alteration of leaf and flower morphology. The role of BBM in promoting cell proliferation and morphogenesis during embryogenesis seems to be confirmed for other species such as Brassica napus and Elaeis guineensis[17, 73]. The AP2 gene is involved in the control of Arabidopsis flower and seed development [68]. This gene is known to be expressed in non-floral organs, such as leaves and stems, and may play a general role in controlling Arabidopsis development. Nevertheless, early activation of these genes, especially HbAP2-12 and HbAP2-18, in proliferating Hevea callus might be not appropriate since it is not conducive to further induction of somatic embryogenesis.
Change occurring during somatic embryogenesis induction
Somatic embryogenesis is triggered by reducing the concentration of growth regulators in the culture medium of Hevea calli. This helps to slow down callus growth to the benefit of embryo formation [74]. Embryogenesis induction is also accompanied by callus browning. In this study, that transition was linked to changes in gene expression. Several ethylene biosynthesis genes, such as HbACS3 and HbACO2, were highly transcribed in the calli before and after embryogenesis induction. When calli were transferred to the EXP medium, transcripts of the SAMS gene accumulated dramatically. That gene is also a marker that differentiated between the regenerant line and the other two low- or non-embryogenic lines, be it for the callus proliferation phase or during somatic embryogenesis induction. SAMS catalyses the formation of S-adenosyl methionine, which is a substrate for the ethylene and polyamine biosynthesis pathways. The latter would seem to play a decisive role in the somatic embryogenesis of Hevea[75]. However, the induction of several ERFs indicates the establishment of ethylene signalling. Four ERFs (HbERF-IVa1, HbERF-Vb2, HbERF-Vb3, HbERF-VI2) and the HbRAV4 gene were transiently induced in calli on the EXP culture medium for somatic embryogenesis induction (Figure 6). At that stage of the process, the comparison between the regenerant line and the low- or non-embryogenic lines also revealed strong induction of 13 ERFs (HbERF-IIIa1, HbERF-IIIb2, HbERF-IIIc1, HbERF-IIIe3, HbERF-IVa1, HbERF-IVa2, HbERF-IVa3, HbERF-Vb2, HbERF-VI-L1, HbERF-VIIa17, HbERF-VIIIa7, HbERF-IXb2, HbERF-IXc4), 2 AP2 (HbAP2-3, HbAP2-7) and the RAV3 gene (Figure 4 or Table 2).
Several of these gene expression markers were predicted to be orthologs of Arabidopsis genes with characterized functions. HbERF-IIIc1 is orthologous to several Arabidopsis DREB1s from group 1 induced by cold [52, 53]. HbERF-IIIe3 is the putative ortholog of TINY known to be activated by drought, cold, ethylene and, to a lesser degree, methyl jasmonate [47]. The semi-dominant tiny mutation causes a reduction in plant height, and affects hypocotyl elongation and fertility. TINY might play a role in communication between biotic and abiotic stress signalling pathways. HbERF-IVa1 and HbERF-IVa2 are two potential orthologs of DREB2A involved in drought-responsive gene expression [50]. HbERF-IVa1 was transitionally induced in calli placed on somatic embryogenesis induction medium, and both HbERF-IVa1 and HbERF-IVa2 were good markers of the regeneration potential compared with the low-embryogenic or non-embryogenic line. These two genes could therefore be very good gene expression markers but would also seem to play a key role in the somatic embryogenesis process. The transcripts of HbERF-VI1 were accumulated in the normal embryos while HbERF-VI2 showed under-expression in the proliferating calli of the regenerant line. These genes are putative orthologs to CRF2, which has been previously described as TARGET OF MP3 (TMO3). The TMO gene is targeted by the auxin-dependent transcription factor MONOPTEROS (MP), which is a regulatory signal in embryonic root specification [59]. AP2 genes are generally transcribed in multiple tissues during development. AILs also play a role in the specification of meristematic or division-competent states [76]. HbAP2-3 is the putative ortholog of two Arabidopsis genes, AIL7/PLT7[66] and AIL6/PLT3, involved in floral meristem growth [67]. HbAP2-7 would seem to be orthologous to the ANT gene [19]. ANT regulates cell proliferation and organ growth by maintaining the meristematic competence of cells during organogenesis [77]. More recently, it was shown to be promoting the initiation and growth of lateral organ primordia, and organ polarity [64]. Consequently, its very high expression in regenerant Hevea callus lines, compared with non-embryogenic and embryogenic lines, was in line with its role in embryo development. With regard to RAV, this family is regulated by ethylene [78] and brassinosteroids [79]. RAVs are involved in the response to biotic and abiotic stress [80]. The increase in HbRAV4 transcripts during somatic embryogenesis induction is in accordance with dramatic changes provoked by ethylene.
Control of development and of somatic embryo quality
The ontogenesis of somatic embryos involves an embryo growth phase, followed by the formation of apical meristem and roots, along with the procambial bundles, and lastly the accumulation of reserves needed for germination [81]. Somatic embryos gradually become dehydrated to acquire the quiescent state [82]. The analysis of AP2/ERF gene expression carried out at the end of the somatic embryo maturation phase revealed that the relative transcript abundance was very high for a large number of genes involved in ethylene biosynthesis and signalling, along with several AP2 genes, including HbAP2-3, HbAP2-6, HbAP2-9, HbAP2-10, HbAP2-13, HbAP2-18 and HbAP2-20 (Figure 5). The potential roles previously described for HbAP2-3 (orthologous to AIL6 and AIL7) and for HbAP2-18 (orthologous to APETALA2) highlight the importance of these genes. All the 16 marker genes discriminating between the normal and abnormal embryos belonged to the ERF family. The abnormal embryos accumulated the transcripts of 6 ERFs belonging to group I (DREB subfamily [50]) more than the normal embryos did. The abundance of the transcripts of another DREB gene, HbERF-IVa2 orthologous to DREB2A, in the normal embryos is worth noting. This suggests differential regulation between the two types of embryos for the ERFs involved in the response to dehydration, salinity and cold.
Conclusion
Of the 132 AP2/ERF genes studied, 40 were expression markers linked to the different stages of the somatic embryogenesis process in Hevea. With the identification of 11 very early markers, it was possible to predict the regeneration potential of proliferating callus lines, which opens up prospects for their application in selecting lines of interest for large-scale propagation. The phylogenetic analysis made it possible to predict more precisely the function of certain genes characterized already in Arabidopsis. The functions of 9 markers suggested that the regulation of hormone and stress signals play just as important a role in somatic embryogenesis as the genes involved in morphogenesis regulation. In addition to these marker genes, another ERF gene expressed during somatic embryogenesis is a potential ortholog of CRF (CYTOKININ RESPONSE FACTOR) encoded by HbERF-VI1 or HbERF-VI2[58]. An in-depth functional characterization of these markers should lead to a better understanding of somatic embryogenesis and explain the loss of embryogenic capacity, and embryo abnormality. Genetic variability in these genes could also be studied to determine whether allelic variations can be used in breeding programmes to select Hevea clones not only for agronomic traits but also for their responsiveness to somatic embryogenesis. The AP2/ERF superfamily could thus play a major role for several other biological functions in Hevea. Firstly, rubber production is stimulated by applying ethephon. In cases of over-tapping in situ coagulation of rubber particles leads to production losses: this is tapping panel dryness. Secondly, the ERF1, ERF2, ERF3 and RAV1 genes, corresponding to HbERF-VIIa1, HbERF-VIIa3, HbERF-VIIa17 and HbRAV1[35], are induced at the same time as secondary laticifer differentiation [83]. Studying this superfamily in Hevea thus provides some new biological knowledge.
Methods
Plant material
The plant regeneration procedure from somatic embryogenesis consists of four steps described in Figure 1. Firstly, primary somatic embryogenesis was carried out using the inner integument of immature seeds from the Hevea clone PB 260 [24]. Secondly, friable callus lines were established from fragments of somatic embryos [29]. Thirdly, callus lines were cryopreserved for long-term storage and to avoid long-term subculturing [33, 39]. Fourthly, embryo development and plant regeneration were induced from friable callus after thawing treatments [24].
In this study, three callus lines with different embryogenic capacity were previously identified from cryopreserved material [33]. The non-embryogenic callus line CI04115 does not have the ability to produce somatic embryos. The embryogenic but non-regenerant callus line CI04106 can produce a few somatic embryos but cannot regenerate plantlets. The regenerant callus line CI07060 can regenerate embryos and plantlets. These cryopreserved callus lines were thawed by immersing the cryovials in a warm water-bath maintained at 37°C for 2 min. ENT medium was prepared with the basic MH macro and microelements supplemented with 9 mM CaCl2, 234 mM sucrose, 30 μM AgNO3, 1.34 μM BAP, 1.34 μM 3,4-D, 0.5 μM ABA and 2.3 g.L-1 Phytagel [24]. The content of the cryovials was placed for 1 h in Petri dishes containing 25 mL of ENT post-thawing medium, which is a modified ENT medium with 1 M sucrose and 1 mM CaCl2[33]. Calli were then transferred for 1 day to fresh ENT post-thawing 2 medium supplemented with 0.5 M sucrose, after which they were transferred again to ENT recovery medium containing a normal sucrose concentration (234 mM) and 1 mM CaCl2. After callus growth recovery in Petri dishes, friable callus aggregates were isolated and transferred to test tubes containing ENT medium. Calli were grown in the dark at 27°C and subcultured regularly every two weeks on fresh ENT medium.
For embryo induction, one gram of friable callus proliferating on ENT medium was transferred to a 250-mL bottle containing 50 mL of expression medium (EXP). EXP was a modified ENT medium supplemented with 9 mM CaCl2, 58.5 mM sucrose, 175.5 mM maltose, 0.44 μM BAP, 0.44 μM 3,4-D and 0.5 μM ABA. This culture was incubated in the dark at 27°C for 4 weeks. Development of somatic embryos was obtained by transferring callus from one bottle to a temporary immersion system containing 200 mL of liquid DEV medium (RITA®, CIRAD, France). There were two successive subcultures of 4 weeks with 1 min of immersion per day in DEV medium, which had the same composition. The DEV medium was a modified ENT medium supplemented with 3 mM CaCl2 and without plant growth regulators. After 8 weeks of culture, mature embryos with a well-formed embryonic axis and cotyledons (normal) were transferred to glass tubes containing a germination medium (GER), which contained MS macro-elements, MH micro-elements and vitamins, and 234 mM sucrose, semi-solidified with 7 g/L Agar. Embryos were cultured under a light intensity of 60 μmol m-2 s-1 with a 12 h day/12 h dark photoperiod for embryo conversion into plantlets. Abnormal embryos were counted and discarded at the end of DEV2.
Total RNA isolation
For the callus lines with different embryogenic capacities, callus was collected at the end of the ENT and EXP cultures. For the regenerant callus line, normal and abnormal embryos were collected at the end of DEV2. Leaves, stems and roots were collected from one-month-old plantlets. All samples were frozen in liquid nitrogen and stored in the freezer at -80°C pending total RNA extraction. Total RNAs were isolated using the caesium chloride cushion method adapted from Sambrook and coll. [84] by Duan and coll. [38]. One gram of fresh matter was ground and transferred to a tube containing 30 mL of extraction buffer consisting of 4 M guanidium isothiocyanate, 1% sarcosine, 1% polyvinylpyrrolidone and 1% ß-mercapto-ethanol. After homogenization, tubes were kept on ice and then centrifuged at 10,000 g at 4°C for 30 min. The supernatant was transferred to a new tube containing 8 mL of 5.7 M CsCl. Ultracentrifugation in a swinging bucket was carried out at 89,705 g at 20°C for 20 h. The supernatant and caesium cushion were discarded whilst the RNA pellet was washed with 70% ethanol. After 30 min of air drying, the pellet was dissolved in 200 μL of sterile water. Although DNA could not cross the caesium cushion for this centrifugation condition, DNA contamination was checked by PCR amplification using primers of the Actin gene including the intron sequence. RNAs were conserved at -80°C.
Primer design and analysis of transcript abundances by real-time RT-PCR
Several rules were applied in order to reduce the risk of error in relative gene expression data. The integrity of total RNA was checked by electrophoresis. Primers were designed at the 3’ side of each sequence in order to reduce the risk of error due to short cDNA synthesis using the Primer 3 module of Geneious (Biomatters Ltd., New Zealand). Real-time PCR amplification and the fusion curve were carried out using a mix of cDNAs in order to check the specificity of each pair of primers. Primer sequences are listed in Additional file 7: Table S1.
cDNAs were synthesized from 2 μg of total RNA to the final 20 μL reaction mixture using a RevertAidTM M-MuLV Reverse Transcriptase (RT) kit according to the manufacturer's instructions (MBI, Fermentas, Canada). Full-length cDNA synthesis was checked on each cDNA sample by PCR amplification of the Actin cDNA using primers at the cDNA ends. Quantitative gene expression analysis was finally carried out by real-time RT-PCR using a Light Cycler 480 (Roche, Switzerland). Real-time PCR reaction mixtures consisted of 2 μL RT product cDNA, 0.6 μL of 5 μM of each primer, and 3 μL 2 × SYBR green PCR master mix (LightCycler® 480 SYBR Green I Master, Roche Applied Sciences) in a 6-μL volume. PCR cycling conditions comprised one denaturation cycle at 95°C for 5 min, followed by 45 amplification cycles (95°C for 20 s, 60°C for 15 s, and 72°C for 20s). Expression analysis was performed in a 384-well plate. Samples were loaded using an automation workstation (Biomek NX, Beckman Coulter).
Real-time PCR was carried out for eleven housekeeping genes in order to select the most stable gene as the internal control for all the compared tissues (HbelF1Aa, HbUBC4, HbUBC2b, HbYLS8, HbRH2b, HbRH8, HbUBC2a, HbalphaTub, Hb40S, HbUbi, HbActin) (Additional file 8: Table S2). HbRH2b was selected as the best reference gene due to its stability in tissues from various stages of somatic embryogenesis. The homogeneity of the HbRH2b gene Cp confirmed that it could be used as an internal reference gene (Table 3). The HbRH2b gene was amplified in each reaction plate in parallel with target genes. The transcript abundance level for each gene was relatively quantified by normalization with the transcript abundance of the reference HbRH2b gene. Relative transcript abundance took into account primer efficiencies. All the normalized ratios corresponding to transcript accumulation were calculated automatically by Light Cycler Software version 1.5.0 provided by the manufacturer using the following calculation: Normalized Ratio = Efficiency -Δ(Cp target-Cp RH2b).
Statistical data analyses
Each callus line was maintained in three biological replicates. Morphological data were recorded and calculated per gram of callus transferred to EXP medium. Statistical analysis was performed with an ANOVA followed by a Student Newman-Keuls test. Values with the same letter did not differ significantly at the 0.05 probability level in Table 1.
Real-time PCR reactions were set up with three biological replications. Statistical analysis was performed with an ANOVA after logarithmic transformation of raw data. The ANOVA was followed by a Student Newman-Keuls test when values of relative transcript abundances were compared for each stage of the somatic embryogenesis process between the three callus lines with different embryogenic capacity (regenerant, embryogenic and non-embryogenic lines), the two types of embryos (normal and abnormal), and the tissues from the normal somatic embryogenesis process from callus to plantlets (regenerant callus on ENT and EXP media, normal embryo, leaf, stem and root). Values with the same letter did not differ significantly at the 0.05 probability level.
In Table 2, the level of expression was calculated as the ratio between the relative transcript abundances in callus of the regenerant line/embryogenic line, the regenerant line/non-embryogenic line grown on ENT and EXP media, and in normal/abnormal somatic embryos. It was considered as an up-regulation with a ratio > 1.0, and a down-regulation with a ratio < 1.0. The statistical analysis was carried out from the logarithm of raw data using the two-tailed probability values of the t test. The ratio of the relative transcript abundances with a p-value ≤ 0.05 was adopted as significant for down or up-regulation. Only significant data are discussed in the manuscript.
Phylogenetic analysis of the AP2 domain from AP2/ERF marker genes
A multiple alignment analysis was performed on full-length AP2 domain sequences from Hevea and Arabidopsis for the AP2, ERF (groups III, IV, V, VII, VIII, X) and RAV families (Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: figure S5 and Additional file 6: Figure S6). The full AP2-domain sequences derived from Hevea and Arabidopsis AP2-domain proteins of around 60 amino acids were then aligned using MUSCLE software [85, 86], which uses a progressive multiple alignment method. The alignment was curated by Gblocks software [87], searching for at least 10-amino-acid-long conserved blocks, and the block with 57 amino acids was extracted. This block of 57 amino acids was used to construct the phylogenetic tree using PhyML software [88], which implements a maximum likelihood tree reconstruction method, using the LG+gamma model, starting from a BioNJ tree [89]. A RAP-Green analysis was performed from the initial PhyML tree to improve gene function inferences and predict gene duplications in phylogenetic trees [90]. The final tree was drawn and displayed with the Archaeoptheryx program, and rooted on the branch separating the RAV family from the rest of the tree. Branch supports were computed using the aLRT-SHlike method [91].
References
Mc Cown BH: Recalcitrance of woody and herbaceous prennial plants: dealing with genetic predeterminism. In Vitro Cell Dev Biol-Plant. 2000, 36: 149-154. 10.1007/s11627-000-0030-6.
Benson E: In vitro plant recalcitrance. Do free radicals have a role in plant tissue culture recalcitrance. In Vitro Cell Dev Biol-Plant. 2000, 36: 163-170.
Benson E: In vitro plant recalcitrance: an introduction. In Vitro Cell Dev Biol-Plant. 2000, 36: 141-148. 10.1007/s11627-000-0029-z.
Michaux-Ferrière N, Carron M-P: Histology of early somatic embryogenesis in Hevea brasiliensis: the importance of the timing of subculturing. Plant Cell Tissue and Organ Culture. 1989, 19: 243-256. 10.1007/BF00043351.
Tisserat B, Murashige T: Effects of ethephon, ethylene, and 2,4-dichlorophenoxyacetic Acid on asexual embryogenesis in vitro. Plant Physiol. 1977, 60 (3): 437-439. 10.1104/pp.60.3.437.
Imin N, Goffard N, Nizamidin M, Rolfe BG: Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol. 2008, 8: 110-10.1186/1471-2229-8-110.
Feng BH, Wu B, Zhang CR, Huang X, Chen YF, Huang XL: Cloning and expression of 1-aminocyclopropane-1-carboxylate oxidase cDNA induced by thidiazuron during somatic embryogenesis of alfalfa (Medicago sativa). J Plant Physiol. 2012, 169 (2): 176-182. 10.1016/j.jplph.2011.08.016.
Karami O, Saidi A: The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol Biol Rep. 2010, 37 (5): 2493-2507. 10.1007/s11033-009-9764-3.
Mantiri FR, Kurdyukov S, Lohar DP, Sharopova N, Saeed NA, Wang XD, Vandenbosch KA, Rose RJ: The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 2008, 146 (4): 1622-1636. 10.1104/pp.107.110379.
El Meskaoui A, Tremblay FM: Involvement of ethylene in the maturation of black spruce embryogenic cell lines with different maturation capacities. J Exp Bot. 2001, 52 (357): 761-769.
Kępczyńska E, Ruduś I, Kępczyński J: Abscisic acid and methyl jasmonate as regulators of ethylene biosynthesis during somatic embryogenesis of Medicago sativa L. Acta Physiol Plant. 2009, 31 (6): 1263-1270. 10.1007/s11738-009-0363-7.
Mantiri FR, Kurdyukov S, Chen SK, Rose RJ: The transcription factor MtSERF1 may function as a nexus between stress and development in somatic embryogenesis in Medicago truncatula. Plant Signal Behav. 2008, 3 (7): 498-500. 10.4161/psb.3.7.6049.
Banno H, Ikeda Y, Niu QW, Chua NH: Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell. 2001, 13 (12): 2609-2618.
Kirch T, Simon R, Grunewald M, Werr W: The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem ccll fate and lateral organ development. Plant Cell. 2003, 15 (3): 694-705. 10.1105/tpc.009480.
Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH: The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006, 47 (11): 1443-1456. 10.1093/pcp/pcl023.
Lau S, Slane D, Herud O, Kong J, Jurgens G: Early embryogenesis in flowering plants: setting up the basic body pattern. Annu Rev Plant Biol. 2012, 63 (1): 483-506. 10.1146/annurev-arplant-042811-105507.
El Ouakfaoui S, Schnell J, Abdeen A, Colville A, Labbe H, Han S, Baum B, Laberge S, Miki B: Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol Biol. 2010, 74 (4-5): 313-326. 10.1007/s11103-010-9674-8.
Boutilier K, Offringa R: Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002, 14: 1737-1749. 10.1105/tpc.001941.
Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR: AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996, 8 (2): 155-168.
Cernac A, Andre C, Hoffmann-Benning S, Benning C: WRI1 is required for seed germination and seedling establishment. Plant Physiol. 2006, 141 (2): 745-757. 10.1104/pp.106.079574.
Cernac A, Benning C: WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40 (4): 575-585. 10.1111/j.1365-313X.2004.02235.x.
Mudunkothge JS, Krizek BA: Three Arabidopsis AIL/PLT genes act in combination to regulate shoot apical meristem function. Plant J. 2012, 71 (1): 108-121. 10.1111/j.1365-313X.2012.04975.x.
Montoro P, Carron MP, Granet F, Lardet L, Leclercq J: Development of new varietal types based on rejuvenation by somatic embryogenesis and propagation by conventional budding or microcutting in Hevea brasiliensis. Acta Horticulturae. 2012, 961: 547.
Carron M-P, Etienne H, Michaux-Ferrière N, Montoro P: Somatic embryogenesis in rubber tree (Hevea brasiliensis Muell. Arg.). Somatic embryogenesis and synthetic seed 1, Volume 30. Edited by: Bajaj YPS. 1995, Berlin, Germany: Springer, 353-369.
Carron M-P, d’Auzac J, Etienne H, El Hadrami I, Housti F, Michaux-Ferrière N, Montoro P: Biochemical and histological features of somatic embryogenesis in Hevea brasiliensis. Indian J Natural Rubber Research. 1992, 5: 7-17.
Wang Z, Zeng C, Chen C, Wu H, Li Q, Fan G, Lu W: Induction of rubber plantlets from anther of Hevea brasiliensis. Chinese J Tropical Crops. 1980, 1: 25-26.
Chen Z: Rubber (Hevea). Handbook of Plant Cell Culture - Crop Species, Volume 2. Edited by: Sharp E. 1984, Hants, England: Macmillan Publishing Company, 546-571.
Carron M-P, Enjalric F: Embryogenese somatique à partir du tégument interne de la graine d'Hevea brasiliensis (Kunth., Muell. Arg.). Comptes Rendus des Séances de l'Académie des Sciences. 1985, 300 (17): 653-658.
Lardet L, Dessailly F, Carron MP, Rio MA, Ferriere N, Montoro P: Secondary somatic embryogenesis in Hevea brasiliensis (Mull. Arg.): an alternative process for long-term somatic embryogenesis. J Rubber Research. 2009, 12 (4): 215-228.
Auboiron E, Carron MP, Michaux-Ferrière N: Influence of atmospheric gases, particularly ethylene, on somatic embryogenesis of Hevea brasiliensis. IRCA/CIRAD Montpellier. 1990, 21: 31-37.
Carron MP, Lardet L, Granet F, Julien J, Teerawatanasuk K, Keli J, Dea BG, Leconte A, Montoro P: Field trials network emphasizes the improvement of growth and yield through micropropagation in rubber tree (Hevea brasiliensis Müll. ARG.). Acta Horticulturae. 2009, 812: 485-492.
Lardet L, Dessailly F, Carron MP, Montoro P, Monteuuis O: Influences of aging and cloning methods on the capacity for somatic embryogenesis of a mature Hevea brasiliensis genotype. Tree Physiol. 2009, 29 (2): 291-298.
Lardet L, Martin F, Dessailly F, Carron MP, Montoro P: Effect of exogenous calcium on post-thaw growth recovery and subsequent plant regeneration of cryopreserved embryogenic calli of Hevea brasiliensis (Mull. Arg.). Plant Cell Rep. 2007, 26 (5): 559-569. 10.1007/s00299-006-0278-3.
Charbit E, Legavre T, Lardet L, Bourgeois E, Ferrière N, Carron MP: Identification of differentially expressed cDNA sequences and histological characteristics of #Hevea brasiliensis# calli in relation to their embryogenic and regenerative capacities. Plant Cell Rep. 2004, 22: 539-548. 10.1007/s00299-003-0737-z.
Duan C, Argout X, Gébelin V, Summo M, Dufayard JF, Leclercq J, Rio M, Champion A, Montoro P: Identification of the Hevea brasiliensis AP2/ERF superfamily in latex. BMC Genomics. 2013, 2013: 2013-in press.
Putranto R, Sanier C, Leclercq J, Duan C, Rio M, Sabau X, Argout X, Montoro P: Differential spatial gene expression in different types of Hevea brasiliensis roots. Plant Sci. 2012, 183: 149-158.
Kuswanhadi , Leclercq J, Rio M, Tregear J, Ducamp-Collin MN, Montoro P: Isolation of three members of the multigene family encoding ACC oxidases in Hevea brasiliensis and investigation of their responses to ethylene stimulation and wounding. J Rubber Research. 2010, 13 (3): 185-205.
Duan C, Rio M, Leclercq J, Bonnot F, Oliver G, Montoro P: Gene expression pattern in response to wounding, methyl jasmonate and ethylene in the bark of Hevea brasiliensis. Tree Physiol. 2010, 30 (10): 1349-1358. 10.1093/treephys/tpq066.
Engelmann F, Lartaud M, Chabrillange N, Carron M-P, Etienne H: Cryopreservation of embryogenic calluses of two commercial clones of Hevea brasiliensis. Cryo Letters. 1997, 18: 107-116.
Wang A, Tan D, Takahashi A, Li T, Harada T: MdERFs, two ethylene-response factors involved in apple fruit ripening. J Exp Bot. 2007, 58: 3743-3748. 10.1093/jxb/erm224.
Babula D, Misztal L, Jakubowicz M, Kaczmarek M, Nowak W, Sadowski J: Genes involved in biosynthesis and signalisation of ethylene in Brassica oleraceaand Arabidopsis thaliana: identification and genome comparative mapping of specific gene homologues. TAG Theoretical and Applied Genetics. 2006, 112 (3): 410-420. 10.1007/s00122-005-0136-7.
Ecker J: The ethylene signal transduction pathway in plants. Science. 1995, 268: 667-675. 10.1126/science.7732375.
Agarwal P, Agarwal P, Reddy M, Sopory S: Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25 (12): 1263-1274. 10.1007/s00299-006-0204-8.
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura H, Yamaguchi-Shinozaki K, Shinozaki K: Two Transcription Factors, DREB1 and DREB2, with an EREBP/AP2 DNA Binding Domain Separate Two Cellular Signal Transduction Pathways in Drought- and LowTemperature-Responsive Gene Expression, Respectively, in Arabidopsis. Plant Cell. 1998, 10: 1391-1406.
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K: AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012, 1819 (2): 86-96. 10.1016/j.bbagrm.2011.08.004.
Cheng MC, Hsieh EJ, Chen JH, Chen HY, Lin TP: Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 2012, 158 (1): 363-375. 10.1104/pp.111.189738.
Wilson K, Long D, Swinburne J, Coupland G: A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell. 1996, 8 (4): 659-671.
Sun S, Yu J-P, Chen F, Zhao T-J, Fang X-H, Li Y-Q, Sui S-F: TINY, a Dehydration-responsive Element (DRE)-binding Protein-like Transcription Factor Connecting the DRE- and Ethylene-responsive Element-mediated Signaling Pathways in Arabidopsis. J Biol Chem. 2008, 283 (10): 6261-6271. 10.1074/jbc.M706800200.
Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M: Arabidopsis Ethylene-Responsive Element Binding Factors Act as Transcriptional Activators or Repressors of GCC Box–Mediated Gene Expression. The Plant Cell Online. 2000, 12 (3): 393-404.
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K: Functional analysis of an arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell Online. 2006, 18 (5): 1292-1309. 10.1105/tpc.105.035881.
Lim CJ, Hwang JE, Chen H, Hong JK, Yang KA, Choi MS, Lee KO, Chung WS, Lee SY, Lim CO: Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem Biophys Res Commun. 2007, 362 (2): 431-436. 10.1016/j.bbrc.2007.08.007.
Novillo F, Medina J, Salinas J: Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci. 2007, 104 (52): 21002-21007. 10.1073/pnas.0705639105.
Gilmour SJ, Fowler SG, Thomashow MF: Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004, 54 (5): 767-781.
Haake V, Cook D, Riechmann J, Pineda O, Thomashow MF, Zhang JZ: Transcription Factor CBF4 Is a Regulator of Drought Adaptation in Arabidopsis. Plant Physiol. 2002, 130 (2): 639-648. 10.1104/pp.006478.
Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K: The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56 (4): 613-626. 10.1111/j.1365-313X.2008.03627.x.
Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K: Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol. 2000, 42 (4): 657-665. 10.1023/A:1006321900483.
Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R: Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153 (2): 757-772. 10.1104/pp.110.155077.
Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ: A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci. 2006, 103 (29): 11081-11085. 10.1073/pnas.0602038103.
Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jurgens G, Weijers D: MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010, 464 (7290): 913-916. 10.1038/nature08836.
Xu Z-S, Chen M, Li L-C, Ma Y-Z: Functions of the ERF transcription factor family in plants. Botany. 2008, 86 (9): 969-977. 10.1139/B08-041.
Ohme-Takagi M, Shinshi H: Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995, 7: 173-182.
Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K: Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005, 58 (4): 585-596. 10.1007/s11103-005-7294-5.
Khandelwal A, Elvitigala T, Ghosh B, Quatrano RS: Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol. 2008, 148 (4): 2050-2058. 10.1104/pp.108.128488.
Nole-Wilson S, Krizek BA: AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 2006, 141 (3): 977-987. 10.1104/pp.106.076604.
Passarinho P, Ketelaar T: BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol. 2008, 68: 225-237. 10.1007/s11103-008-9364-y.
Prasad K, Grigg SP, Barkoulas M, Yadav RK, Sanchez-Perez GF, Pinon V, Blilou I, Hofhuis H, Dhonukshe P, Galinha C, et al: Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr Biol. 2011, 21 (13): 1123-1128. 10.1016/j.cub.2011.05.009.
Krizek BA: Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family. J Exp Bot. 2011, 62 (10): 3311-3319. 10.1093/jxb/err127.
Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK: Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994, 6 (9): 1211-1225.
Wurschum T, Gross-Hardt R, Laux T: APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell. 2006, 18 (2): 295-307. 10.1105/tpc.105.038398.
Woo HR, Kim JH, Kim J, Kim J, Lee U, Song I-J, Kim J-H, Lee H-Y, Nam HG, Lim PO: The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot. 2010, 61 (14): 3947-3957. 10.1093/jxb/erq206.
Michaux-Ferrière N, Carron MP: Histological structure of calli and induction of somatic embryos in Hevea brasiliensis according to subculture sequences. ompte Rendu du Colloque Exploitation, Physiologie et Amélioration de l'Hevea: 2-7. 1988, CARRON: IRCA, 479-490. November 1988
Srinivasan C, Liu Z, Heidmann I, Supena ED, Fukuoka H, Joosen R, Lambalk J, Angenent G, Scorza R, Custers JB, et al: Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta. 2007, 225 (2): 341-351.
Morcillo F, Gallard A, Pillot M, Jouannic S, Aberlenc-Bertossi F, Collin M, Verdeil JL, Tregear JW: EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta. 2007, 226 (6): 1353-1362. 10.1007/s00425-007-0574-3.
Blanc G, Michaux-Ferrière N, Teisson C, Lardet L, Carron MP: Effects of carbohydrate addition on the induction of somatic embryogenesis in Hevea brasiliensis. Plant Cell Tissue and Organ Culture. 1999, 59: 103-112. 10.1023/A:1006437731011.
El Hadrami I, Michaux-Ferrière N, Carron MP, D'Auzac J: Polyamines (PA.), a possible limiting factor in somatic embryogenesis of Hevea brasiliensis. CR Acad Sci Paris. 1989, 308 (Série III): 205-211.
Nole-Wilson S, Tranby TL, Krizek BA: AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol. 2005, 57 (5): 613-628. 10.1007/s11103-005-0955-6.
Mizukami Y, Fischer RL: Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000, 97 (2): 942-947. 10.1073/pnas.97.2.942.
Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR: Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA. 2003, 100 (5): 2992-2997. 10.1073/pnas.0438070100.
Hu YX, Wang YH, Liu XF, Li JY: Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004, 14 (1): 8-15. 10.1038/sj.cr.7290197.
Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK: Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol. 2006, 61 (6): 897-915. 10.1007/s11103-006-0057-0.
Michaux Ferrière N, Grout H, Carron MP: Origin and ontogenesis of somatic embryos in Hevea brasiliensis. Am J Bot. 1992, 79 (2): 174-180. 10.2307/2445105.
Etienne H, Montoro P, Michaux-Ferrière N, Carron MP: Effects of desiccation, medium osmolarity and abscisic acid on the maturation of Hevea brasiliensis somatic embryos. J Exp Bot. 1993, 44 (267): 1613-1619.
Wu H, Yu B, Cheng Q, Zeng R, Duan C, Nie Z, Li Y: Cloning and Characterization of Jasmonic Acid-Induced AP2/EREBP Genes in Laticifer from Rubber Tree (Hevea brasiliensis Muell. Arg.). Chinese Agricultural, Science Bulletin. 2010, 2010 (5): 287-293.
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning, a laboratory manual. 1989, CHS Press.
Edgar RC: MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004, 5: 113-10.1186/1471-2105-5-113.
Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32 (5): 1792-1797. 10.1093/nar/gkh340.
Talavera G, Castresana J: Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007, 56 (4): 564-577. 10.1080/10635150701472164.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O: New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010, 59 (3): 307-321. 10.1093/sysbio/syq010.
Gascuel O: BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997, 14 (7): 685-695. 10.1093/oxfordjournals.molbev.a025808.
Dufayard JF, Duret L, Penel S, Gouy M, Rechenmann F, Perriere G: Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005, 21 (11): 2596-2603. 10.1093/bioinformatics/bti325.
Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O: Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 2011, 60 (5): 685-699. 10.1093/sysbio/syr041.
Acknowledgements
This work was supported by the Institut Français du Caoutchouc and a fellowship of the Rubber Research Institute of Thailand. The authors thank Peter Biggins for the English version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors’ contributions
FD and FM established the various friable callus lines for this experiment from cryopreserved material. PP induced somatic embryogenesis and regenerated both embryos and plantlets, and then carried out the total RNA isolation and cDNA synthesis from samples. PP and RP designed the primers from the AP2/ERF genes. MR and JL supervised the real-time RT-PCR analysis on the Roche platform. LL supervised the tissue culture experiments. JFD advised on the strategy and supervised the phylogenetic analyses. PP and PM drafted the manuscript. PM coordinated this research work. All the authors read and approved the final manuscript.
Electronic supplementary material
12870_2012_1209_MOESM1_ESM.pdf
Additional file 1: Figure S1: Phylogenetic tree of the AP2 family. The deduced amino acid sequences of the AP2 domain from Hevea (black letter) and Arabidopsis (blue letter) were aligned using Muscle, and the phylogenetic tree was constructed using PhyML with an LG+T model. Hevea somatic embryogenesis marker genes are indicated in bold letters. (PDF 154 KB)
12870_2012_1209_MOESM2_ESM.pdf
Additional file 2: Figure S2: Phylogenetic tree of ERF group I. The deduced amino acid sequences of the AP2 domain from Hevea (black letter) and Arabidopsis (blue letter) were aligned using Muscle, and the phylogenetic tree was constructed using PhyML with an LG+T model. Hevea somatic embryogenesis marker genes are indicated in bold letters. (PDF 91 KB)
12870_2012_1209_MOESM3_ESM.pdf
Additional file 3: Figure S3: Phylogenetic tree of ERF group III. The deduced amino acid sequences of the AP2 domain from Hevea (black letter) and Arabidopsis (blue letter) were aligned using Muscle, and the phylogenetic tree was constructed using PhyML with an LG+T model. Hevea somatic embryogenesis marker genes are indicated in bold letters. (PDF 161 KB)
12870_2012_1209_MOESM4_ESM.pdf
Additional file 4: Figure S4: Phylogenetic tree of ERF group IV. The deduced amino acid sequences of the AP2 domain from Hevea (black letter) and Arabidopsis (blue letter) were aligned using Muscle, and the phylogenetic tree was constructed using PhyML with an LG+T model. Hevea somatic embryogenesis marker genes are indicated in bold letters. (PDF 72 KB)
12870_2012_1209_MOESM5_ESM.pdf
Additional file 5: Figure S5: Phylogenetic tree of ERF group VI. The deduced amino acid sequences of the AP2 domain from Hevea (black letter) and Arabidopsis (blue letter) were aligned using Muscle, and the phylogenetic tree was constructed using PhyML with an LG+T model. Hevea somatic embryogenesis marker genes are indicated in bold letters. (PDF 62 KB)
12870_2012_1209_MOESM6_ESM.pdf
Additional file 6: Figure S6: Phylogenetic tree of ERF group X. The deduced amino acid sequences of the AP2 domain from Hevea (black letter) and Arabidopsis (blue letter) were aligned using Muscle, and the phylogenetic tree was constructed using PhyML with an LG+T model. Hevea somatic embryogenesis marker genes are indicated in bold letters. (PDF 83 KB)
12870_2012_1209_MOESM7_ESM.xlsx
Additional file 7: Table S1: List of primer sequences for 132 AP2/ERF genes from H. brasiliensis clone PB 260. (XLSX 19 KB)
12870_2012_1209_MOESM8_ESM.xlsx
Additional file 8: Table S2: Comparison Cp value of callus with various embryogenic capacities grown on embryogenesis induction medium (ENT) and somatic embryogenesis expression medium (EXP){Calli are from three types of lines: (NE) non-embryogenic line (CI04115), (E) embryogenic and non-regenerant line (CI04106), and (R) regenerant line (CI07060)}, normal and abnormal embryo and 3 organ of plantlet:(L) Leaf, (S) Stem, (R) Root in 11 house keeping genes. (XLSX 14 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Piyatrakul, P., Putranto, RA., Martin, F. et al. Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis. BMC Plant Biol 12, 244 (2012). https://doi.org/10.1186/1471-2229-12-244
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-12-244