Abstract
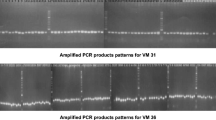
The genetic diversity of cowpea (Vigna unguiculata L. Walp.) in Ethiopia was analyzed using 19 uniform accessions, 62 variable accessions (yielding 185 sub-types), and two mungbean (Vigna radiata) accessions (four subtypes) as outgroup. A set of 23 polymorphic simple sequence repeat (SSR) markers was identified, and polymorphism in the various accessions was scored by determining amplicon variability. Allele frequency, genetic diversity, and polymorphism information content (PIC) were determined for each SSR marker, and a neighbor joining dendrogram was generated to show the genetic relationship among the individual accessions. A total of 75 allelic variants was defined, with the average number of alleles per locus calculated to be three. The average genetic diversity (D) was 0.47, and PIC was 0.4. Three main clusters were identified by phylogenetic analysis, and the clusters and sub-grouping were supported by STRUCTURE and principal component analysis. This grouping had a moderate fixation index value of 0.075 and gene flow (Nm) of 3.176, indicating that the accessions possess wide diversity within individuals and populations. The accessions showed no clustering by geographical origins. Three well-characterized molecular markers (SSR1, C42-2B, and 61RM2) for race specific resistance to Striga gesnerioides in the cowpea cultivar B301 were used to evaluate the accessions for their potential for use in genetic improvement against this pest. Based on this analysis, only two accessions, 222890–2 from Gambela and 286–2 from the Southern Nations, Nationalities, and Peoples (SNNP) region, were found to cluster with B301 and contain the SSR1 resistance allele. These findings will assist in germplasm conservation efforts by the Institute of Biodiversity and Conservation of Ethiopia, and contribute to future studies aimed at the genetic improvement of local germplasm for improved overall agronomic performance as well as Striga resistance in particular.





Similar content being viewed by others
References
Abe J, Xu D, Suzuki Y, Kanazawa A, Shimamoto Y (2003) Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor Appl Genet 106:445–453
Andargie M, Pasquet RS, Gowda BS, Muluvi GM, Timko MP (2011) Construction of a SSR-based genetic map and identification of QTL for domestication traits using recombinant inbred lines from a cross between wild and cultivated cowpea (V. unguiculata (L.) Walp.). Mol Breed 28:413–420
Asare AT, Gowda BS, Galyuon IK, Aboagye LL, Takrama JF, Timko MP (2010) Assessment of the genetic diversity in cowpea (Vigna unguiculata L. Walp.) germplasm from Ghana using simple sequence repeat markers. Plant Genet Res 8:142–150
Asare AT, Galyuon IK, Padi FK, Otwe EP, Takrama J (2013) Responses of recombinant inbred lines of cowpea [(Vigna unguiculata (l.) walp] to Striga gesnerioides infestation in Ghana. Eur Sci J 9:503–513
Ba FS, Pasquet RS, Gepts P (2004) Genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] as revealed by RAPD markers. Genet Resour Crop Evol 51:539–550
Badiane FA, Diouf D, Sané D, Diouf O, Goudiaby V, Diallo N (2004) Screening cowpea [Vigna unguiculata (L.) Walp.] varieties by inducing water deficit and RAPD analyses. Afr J Biotechnol 3:174–178
Badiane F, Gowda B, Cissé N, Diouf D, Sadio O, Timko M (2012) Genetic relationship of cowpea (Vigna unguiculata) varieties from Senegal based on SSR markers. Genet Mol Res 11:292–304
Barrett B, Kidwell K (1998) AFLP-based genetic diversity assessment among wheat cultivars from the Pacific Northwest. Crop Sci 38:1261–1271
Botanga CJ, Timko MP (2006) Phenetic relationships among different races of Striga gesnerioides (Willd.) Vatke from West Africa. Genome 49:1351–1365
Choumane W, Winter P, Weigand F, Kahl G (2000) Conservation and variability of sequence-tagged microsatellite sites (STMSs) from chickpea (Cicer aerietinum L.) within the genus Cicer. Theor Appl Genet 101:269–278
De Vicente M, Lopez C, Fulton T (2004) Genetic diversity analysis with molecular marker data: learning module. International Plant Genetic Resources Institute (IPGRI), Cornell University
Dikshit H, Jhang T, Singh N, Koundal K, Bansal K, Chandra N, Tickoo J, Sharma T (2007) Genetic differentiation of Vigna species by RAPD, URP and SSR markers. Biol Plant 51:451–457
Diouf D, Hilu KW (2005) Microsatellites, RAPD markers to study genetic relationships among cowpea breeding lines and local varieties in Senegal. Genet Resour Crop Evol 52:1057–1067
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing structure output and implementing the evanno method. Conserv Genet Resour 4:359–361
Ehlers J, Hall A (1996) Genotypic classification of cowpea based on responses to heat and photoperiod. Crop Sci 36:673–679
Fang J, Chao C-CT, Roberts PA, Ehlers JD (2007) Genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] in four West African and USA breeding programs as determined by AFLP analysis. Genet Resour Crop Evol 54:1197–1209
Fatokun C, Danesh D, Young N, Stewart E (1993) Molecular taxonomic relationships in the genus Vigna based on RFLP analysis. Theor Appl Genet 86:97–104
Fotso M, Azanza J-L, Pasquet R, Raymond J (1994) Molecular heterogeneity of cowpea (Vigna unguiculata, Fabaceae) seed storage proteins. Plant Syst Evol 191:39–56
Gemechu K, Ali K, Makkouk K, Halila M, Malhotra R, Ahmed S, Surenda B (2003) Food and forage legumes of Ethiopia: progress and prospects. Proceedings of the 13th Annual Research and Extension Review Meeting, Addis Ababa, Ethiopia, pp. 124–130
He C, Poysa V, Yu K (2003) Development and characterization of simple sequence repeat (SSR) markers and their use in determining relationships among Lycopersicon esculentum cultivars. Theor Appl Genet 106:363–373
Hegde V, Mishra S (2009) Landraces of cowpea, Vigna unguiculata (L.) Walp., as potential sources of genes for unique characters in breeding. Genet Resour Crop Evol 56:615–627
Kiambi D, Newbury H, Ford-Lloyd B, Dawson I (2005) Contrasting genetic diversity among Oryza longistaminata (A. Chev et Roehr) populations from different geographic origins using AFLP. Afr J Biotechnol 4:308–317
Kumar MD (2011) Pulse Crop production: principles and technologies. PHI Learning, New Delhi
Kuruma R, Kiplagat O, Ateka E, Ouoche G (2008) Genetic diversity of Kenya cowpea accessions based on morphological and microsatellite markers. East Afr Agric For J 76:3–4
Lane JA, Moore THM, Child DV, Cardwell K (1996) Characterization of virulence and geographic distribution of Striga gesnerioides on cowpea in West Africa. Plant Dis 80:299–301
Lane JA, Moore THM, Child DV, Bailey JA (1997) Variation in virulence of Striga gesnerioides on cowpea: new sources of resistance. In: Singh BB, Mohan Raj DR, Dashiell KE, Jackai LEN (eds) Advances in cowpea research. International Institute for Tropical Agricultural Sciences, Ibadan, pp 225–230
Langyintuo A, Lowenberg-Deboer J, Faye M, Lambert D, Ibro G, Moussa B, Kergna A, Kushwaha S, Musa S, Ntoukam G (2003) Cowpea supply and demand in West and Central Africa. Field Crop Res 82:215–231
Li Wang M, Barkley NA, Gillaspie GA, Pederson GA (2008) Phylogenetic relationships and genetic diversity of the USDA Vigna germplasm collection revealed by gene-derived markers and sequencing. Genet Res 90:467–480
Li J, Timko MP (2009) Gene-for-gene resistance in striga-cowpea associations. Science 325:1094–1094
Li C-D, Fatokun CA, Ubi B, Singh BB, Scoles GJ (2001) Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci 41:189–197
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Nkongolo K (2003) Genetic characterization of Malawian cowpea (Vigna unguiculata (L.) Walp) landraces: diversity and gene flow among accessions. Euphytica 129:219–228
O’Neill R, Snowdon R, Köhler W (2003) Population genetics: aspects of biodiversity. Prog Bot 64:115–137
Ogunkanmi L, Ogundipe O, Ng N, Fatokun C (2008) Genetic diversity in wild relatives of cowpea (Vigna unguiculata) as revealed by simple sequence repeats (SSR) markers. J Food Agric Environ 6:253–268
Pasquet RS (1993) Variation at isozyme loci in wild Vigna unguiculata (Fabaceae, Phaseoleae). Plant Syst Evol 186:157–173
Pasquet R (1999) Genetic relationships among subspecies of Vigna unguiculata (L.) Walp. based on allozyme variation. Theor Appl Genet 98:1104–1119
Pasquet R (2000) Allozyme diversity of cultivated cowpea Vigna unguiculata (L.) Walp. Theor Appl Genet 101:211–219
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLPs. Theor Appl Genet 97:1248–1255
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Enfield, Plymouth, pp 43–76
Plaschke J, Ganal M, Röder M (1995) Detection of genetic diversity in closely related bread wheat using microsatellite markers. Theor Appl Genet 91:1001–1007
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Riahi K, Krey V, Rao S, Chirkov V, Fischer G, Kolp P, Kindermann G, Nakicenovic N, Rafai P (2011) RCP-8.5: exploring the consequence of high emission trajectories. Clim Chang 109:33–57. doi:10.1007/s10584-011-0149-y
Rongwen J, Akkaya M, Bhagwat A, Lavi U, Cregan P (1995) The use of microsatellite DNA markers for soybean genotype identification. Theor Appl Genet 90:43–48
Sawadogo M, Ouedraogo JT, Gowda BS, Timko MP (2010) Genetic diversity of cowpea (Vigna unguiculata L. Walp) cultivars in Burkina Faso resistant to Striga gesnerioides. Afr J Biotechnol 9:8146–8153
Scoles G, Simon M, Benko-Iseppon A-M, Resende L, Winter P, Kahl G (2007) Genetic diversity and phylogenetic relationships in Vigna Savi germplasm revealed by DNA amplification fingerprinting. Genome 50:538–547
Singh B (2005) Cowpea [Vigna unguiculata (L.) Walp.]. In: Singh RJ, Jauhar PP (eds) Genetic resources, chromosome engineering and crop improvement, vol 1. CRC, Boca Raton, pp 117–162
Timko MP, Singh B (2008) Cowpea, a multifunctional legume. In: Moore PH, Ming R (eds) Genomics of tropical crop plants. Springer, New York, pp 227–258
Timko MP, Ehlers JD, Roberts PA (2007) Cowpea. In: Berlin KC (ed) Genome mapping and molecular breeding in plants, pulses, sugar and tuber crops, vol 3. Springer, Berlin, pp 49–68
Tosti N, Negri V (2002) Efficiency of three PCR-based markers in assessing genetic variation among cowpea (Vigna unguiculata subsp. unguiculata) landraces. Genome 45:268–275
Uma M, Hittalamani S, Murthy B, Viswanatha K (2009) Microsatellite DNA marker aided diversity analysis in cowpea [Vigna unguiculata (L.) Walp]. Indian J Genet Plant Breeding 69:35–43
Vaillancourt R, Weeden N (1992) Chloroplast DNA polymorphism suggests Nigerian center of domestication for the cowpea, Vigna unguiculata (Leguminosae). Am J Bot 79:1194–1199
Wright S (1984) Evolution and the genetics of populations, vol 3: experimental results and evolutionary deductions. The University of Chicago Press, London
Xavier GR, Martins LMV, Rumjanek NG, Freire Filho FR (2005) Variabilidade genética em acessos de caupi analisada por meio de marcadores RAPD. Pesq Agrop Brasileira 40:353–359
Xu P, Wu X, Wang B, Liu Y, Qin D, Ehlers JD, Close TJ, Hu T, Lu Z, Li G (2010) Development and polymorphism of (Vigna unguiculata ssp. unguiculata) microsatellite markers used for phylogenetic analysis in asparagus bean (Vigna unguiculata ssp. sesquipedialis (L.) Verdc.). Mol Breed 25:675–684
Yang Z, Goldman N, Friday A (1994) Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol Biol Evol 11:316–324
Zannouou A, Kossou D, Ahanchédé A, Zoundjihékpon J, Agbicodo E, Struik P, Sanni A (2008) Genetic variability of cultivated cowpea in Benin assessed by random amplified polymorphic DNA. Afr J Biotechnol 7:4407–4414
Acknowledgments
The authors wish to thank the members of the Timko lab for their help in various aspects of this work and for helpful suggestions on this manuscript, especially Sory Diallo, Frederick Awuku, Yu Wang and Tatyana Katova. These studies were supported by funds from the Kirkhouse Trust. While the research reported here was funded by the Kirkhouse Trust, the design, execution, and interpretation of the research remains wholly the responsibility of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Desalegne, B.A., Mohammed, S., Dagne, K. et al. Assessment of genetic diversity in Ethiopian cowpea [Vigna unguiculata (L.) Walp.] germplasm using simple sequence repeat markers. Plant Mol Biol Rep 34, 978–992 (2016). https://doi.org/10.1007/s11105-016-0979-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-016-0979-x