Abstract
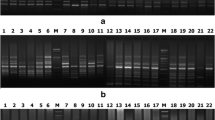
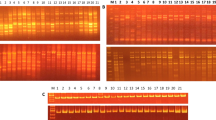
Seventy genotypes belonging to 7 wild and cultivated Vigna species were genetically differentiated using randomly amplified polymorphic DNA (RAPD), universal rice primer (URP) and simple sequence repeat (SSR) markers. We identified RAPD marker, OPG13 which produced a species-specific fingerprint profile. This primer characterized all the Vigna species uniquely suggesting an insight for their co-evolution, domestication and interspecific relationship. The cluster analysis of combined data set of all the markers resulted in five major groups. Most of the genotypes belonging to cultivated species formed a specific group whereas all the wild species formed a separate cluster using unweighted paired group method with arithmetic averages and principle component analysis. The Mantel matrix correspondence test resulted in a high matrix correlation with best fit (r = 0.95) from combined marker data. Comparison of three-marker systems showed that SSR marker was more efficient in detecting genetic variability among all the Vigna species. The narrow genetic base of the V. radiata cultivars obtained in the present study emphasized that large germplasm collection should be used in Vigna improvement programme.
Similar content being viewed by others
Abbreviations
- AFLP:
-
amplified fragment length polymorphism
- CTAB:
-
cetyltrimethyl ammonium bromide
- EMR:
-
effective multiplex ratio
- Hav :
-
arithmetic mean heterozygosity
- Hn:
-
expected heterozygosity
- ISSR:
-
inter simple sequence repeats
- MI:
-
marker index
- PCA:
-
principal components analysis
- PCR:
-
polymerase chain reaction
- RAPD:
-
randomly amplified polymorphic DNA
- Rp:
-
resolving power
- SAHN:
-
sequential agglomerative hierarical nested
- SAMPL:
-
selective amplification of microsatellite polymorphic loci
- SSR:
-
simple sequence repeat
- STMS:
-
sequence tagged microsatellite
- UPGMA:
-
unweighted paired group method with arithmetic averages
- URP:
-
universal rice primer
References
Ajibade, S.R., Weeden, N.F., Chite, S.M.: Inter simple sequence repeat analysis of genetic relationship in genus Vigna.-Euphytica 111: 47–55, 2000.
Banerjee, H., Pai, R.A., Sharma R.P.: Restriction fragment length polymorphism and random amplified polymorphic DNA analysis of chickpea accessions.-Biol. Plant. 42: 197–208, 1999.
Betal, S., Roy Chowdhury, P., Kundu, S., Sen Raychaudhuri, S.: Estimation of genetic variability of Vigna radiata cultivars by RAPD analysis.-Biol. Plant. 48: 205–209, 2004.
Kang, H.W., Park, D.S., Go, S.J., Eun, M.Y.: Fingerprinting of diverse genomes using PCR with universal rice primers generated from repetitive sequences of Korean weedy rice.-Mol. Cells 3: 281–287, 2002.
Lakhanpaul, S., Chadha, S., Bhat, K.V.: Random amplified polymorphic DNA (RAPD) analysis in Indian mungbean (Vigna radiata L. Wilczek) cultivars.-Genetica 109: 227–234, 2000.
Lambridges, C.L., Lawn, R.L.: Two genetic linkage maps of mungbean using RFLP and RAPD markers.-Aust. J. agr. Res. 51: 415–425, 2000.
Lawn, R.J., Williams, R.W., Imrie, B.C.: Potential of wild germplasm as a source of tolerance to environmental stresses in mungbean.-In: Shanmugasundaram, S., McLean, B.T. (ed.): Proceedings of the 2nd International Symposium on Mungbean. Pp. 136–180. AVRDC, Shanva 1987.
Mantel, N.: The deduction of disease clustering and a generalized regression approach.-Cancer Res. 27: 209–220, 1967.
Murray, M.G., Thompson, W.F.: Rapid isolation of high molecular weight plant DNA.-Nucl. Acids Res. 8: 4321–4325, 1980.
Nei, M.: Analysis of gene diversity in subdivided populations.-Proc. nat. Acad. Sci. USA 70: 3321–3323, 1973.
Powell, W., Morgante, M., Andre, C., Hanafey, M., Vogel, J., Tingey, S., Rafalski, A.: The comparison of RFLP, AFLP and SSR (microsatellite) markers for germplasm analysis.-Mol. Breed. 2: 225–238, 1996.
Prevost, A., Wilkinson, M.J.: A new system of comparing PCR primers applied to ISSR finger printing of potato cultivars.-Theor. appl. Genet. 102: 440–449, 1999.
Rohlf, F.J.: NTSYS-PC: Numerical Taxonomy and Multivariate Analysis System. Version 2.11T.-Exeter Software, Setauket 2000.
Samec, P., Pošvec, Z., Stejskal, J., Našinec, V., Griga, M.: Cultivar identification and relationships in Pisum sativum L. based on RAPD and isoenzymes.-Biol. Plant. 41: 39–48, 1998.
Santalla, M., Power, J.B., Davery, M.R.: Genetic diversity in mungbean germplasm revealed by RAPD markers.-Plant Breed. 5: 473–478, 1998.
Singh, K.P., Sareen, M., Ashwini, P.K., Kumar, A.: Interspecific hybridization studies in V. radiata (L.) Wilczek and Vigna umbellata.-Nat. J. Plant Improv. 3: 16–18, 2003
Souframanien, J., Gopalakrishna, T.: A comparative analysis of genetic diversity in blackgram genotypes using RAPD and ISSR markers.-Theor. appl. Genet. 109: 1687–93, 2004.
Srinivas, P., Haulpalai, N., Saengehot, S., Ngampongsai, N.: The use of wild relatives and gamma radiation in mungbean and blackgram.-In: Vaughan, D. (ed.): Wild Legumes. The VII MAAF International workshop on Genetic Resources. Pp. 205–218. NIAB, Tsukuba 1999.
Tosti, N., Negri, V.: Efficiency of three PCR based markers in assessing genetic variation among cowpea (Vigna unguiculata) landraces.-Genome 45: 268–275, 2002.
Wang, X.W., Kaga, A., Tomooka, N., Vaughan, D.A.: The development of SSR markers by a new method in plants and their application to gene flow studies in adzuki bean [Vigna angularis (Willd.) Ohwi & Ohashi].-Theor. appl. Genet. 109: 352–360, 2004.
Willams, J.G.K., Kubelik, A.R., Lavak, K.J., Rafalski, J.A., Tingey, S.V.: DNA polymorphism amplified by arbitrary primers are useful as genetic markers.-Nucl. Acids Res. 18: 6531–6535, 1990.
Yu, K., Park, S.J., Poysa, V., Gepts, P.: Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.)-J. Heredity 91: 429–434, 2000.
Zong, X.X., Kaga, A., Tomooka, N., Wang, X.W., Han, O.K., Vaughan, D.: The genetic diversity of the Vigna angularis complex in Asia.-Genome 46: 647–58, 2003.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dikshit, H.K., Jhang, T., Singh, N.K. et al. Genetic differentiation of Vigna species by RAPD, URP and SSR markers. Biol Plant 51, 451–457 (2007). https://doi.org/10.1007/s10535-007-0095-8
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10535-007-0095-8