Abstract
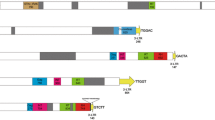
In the study, we developed new markers for phylogenetic relationships and intraspecies differentiation in Coffea. Nana and Divo, two novel Ty1-copia LTR-retrotransposon families, were isolated through C. canephora BAC clone sequencing. Nana- and Divo-based markers were used to test their: (1) ability to resolve recent phylogenetic relationships; (2) efficiency in detecting intra-species differentiation. Sequence-specific amplification polymorphism (SSAP), retrotransposon-microsatellite amplified polymorphism (REMAP) and retrotransposon-based insertion polymorphism (RBIP) approaches were applied to 182 accessions (31 Coffea species and one Psilanthus accession). Nana- and Divo-based markers revealed contrasted transpositional histories. At the BAC clone locus, RBIP results on C. canephora demonstrated that Nana insertion took place prior to C. canephora differentiation, while Divo insertion occurred after differentiation. Combined SSAP and REMAP data showed that Nana could resolve Coffea lineages, while Divo was efficient at a lower taxonomic level. The combined results indicated that the retrotransposon-based markers were useful in highlighting Coffea genetic diversity and the chronological pattern of speciation/differentiation events. Ongoing complete sequencing of the C. canephora genome will soon enable exhaustive identification of LTR-RTN families, as well as more precise in-depth analyses on contributions to genome size variation and Coffea evolution.




Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42(1):251–269
Bousios A, Saldana-Oyarzabala I, Valenzuela-Zapatab GA, Wooda C, Pearce RS (2007) Isolation and characterization of Ty1-copia retrotransposon sequences in the blue agave (Agave tequilana Weber var. azul) and their development as SSAP markers for phylogenetic analysis. Plant Sci 172(2):291–298
Bustamante-Porras J, Campa C, Poncet V, Noirot M, Leroy T, Hamon S, de Kochko A (2007) Molecular characterization of an ethylene receptor gene (CcETR1) in coffee trees, its relationship with fruit development and caffeine content. Mol Genet Genomics 277:701–712
Charrier A (1978) La structure génétiques des caféiers spontanés de la région malgache (Mascarocoffea). Leurs relations avec les caféiers d’origine africaine (Eucoffea). Thèse d’état Université Paris-Sud Orsay, p 221
Chevalier A (1947) Les caféiers du globe. III Systématique des caféiers et faux caféiers. Maladies et insectes nuisibles. Lechevalier P ed Paris
Coulibaly I, Revol B, Noirot M, Poncet V, Lorieux M, Carasco-Lacombe C, Minier J, Dufour M, Hamon P (2003) AFLP and SSR polymorphism in a Coffea interspecific backcross progeny [(C. heterocalyx × C. canephora) × C. canephora]. Theor Appl Genet 107(6):1148–1155
Couturon E, Lashermes P, Charrier A (1998) First intergeneric hybrids (Psilanthus ebracteolatus Hiern × Coffea arabica L.) in coffee trees. Can J Bot 76(3):542–546
Cros J, Combes MC, Trouslot P, Anthony F, Hamon S, Charrier A, Lashermes P (1998) Phylogenetic analysis of chloroplast DNA variation in Coffea L. Mol Phylogenet Evol 9(1):109–117
Cubry P, Musoli P, Legnate H, Pot D, de Bellis F, Poncet V, Anthony F, Dufour M, Leroy T (2008) Diversity in coffee assessed with SSR markers: structure of the genus Coffea and perspectives for breeding. Genome 51(1):50–63
D’Onofrio C, Lorenzis G, Giordani T, Natali L, Cavallini A, Scalabrelli G (2010) Retrotransposon-based molecular markers for grapevine species and cultivars identification. Tree Genet Genomes 6:451–466
Davis AP, Govaerts R, Bridson DM, Stoffelen P (2006) An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot J Linn Soc 142(4):465–512
de Kochko A, Akaffou S, Andrade AC, Campa C, Crouzillat D, Guyot R, Hamon P, Ming R, Mueller LA, Poncet V, Tranchant-Dubreuil C, Hamon S (2010) Advances in coffea genomics. Adv Bot Res 53:23–63
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Dussert S, Lashermes P, Anthony F, Montagnon C, Trouslot P, Combes BC, Berthaud J, Noirot M, Hamon S (1999) Coffee (Coffea canephora). In: Hamon, P Seguin, M Perrier, X Glaszmann, JC (eds) Genetic diversity of cultivated tropical plants. CIRAD, Montpellier, p 376
FAO (ed) (2006) Global Forest Resources Assessment 2005. Progress towards sustainable forest management. FAO Forestry Paper No. 147, Rome
Flavell AJ, Knox MR, Pearce SR, Ellis TH (1998) Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J 16(5):643–650
Gardner TA, Barlow J, Chazdon R, Ewers RM, Harvey CA, Peres CA, Sodhi NS (2009) Prospects for tropical forest biodiversity in a human-modified world. Ecol Lett 12(6):561–582
Gomez C, Dussert S, Hamon P, Hamon S, de Kochko A, Poncet V (2009) Current genetic differentiation of Coffea canephora Pierre ex A. Froehn in the Guineo-Congolian African zone: cumulative impact of ancient climatic changes and recent human activities. BMC Evol Biol 9:167
Guyot R, de la Mare M, Viader V, Hamon P, Coriton O, Bustamante-Porras J, Poncet V, Campa C, Hamon S, de Kochko A (2009) Microcollinearity in an ethylene receptor coding gene region of the Coffea canephora genome is extensively conserved with Vitis vinifera and other distant dicotyledonous sequenced genomes. BMC Plant Biol 9(1):22
Hamon P, Siljak-Yakovlev S, Srisuwan S, Robin O, Poncet V, Hamon S, de Kochko A (2009) Physical mapping of rDNA and heterochromatin in chromosomes of 16 Coffea species: a revised view of species differentiation. Chromosome Res 17(3):291–304
Hill P, Burford D, Martin DM, Flavell AJ (2005) Retrotransposon populations of Vicia species with varying genome size. Mol Genet Genomics 273(5):371–381
Jing R, Vershinin A, Grzebyta J, Shaw P, Smykal P, Marshall D, Ambrose MJ, Ellis TH, Flavell AJ (2010) The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol Biol 10:44
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS (2000) Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97(12):6603–6607
Konovalov FA, Goncharov NP, Goryunova S, Shaturova A, Proshlyakova T, Kudryavtsev A (2010) Molecular markers based on LTR retrotransposons BARE-1 and Jeli uncover different strata of evolutionary relationships in diploid wheats. Mol Genet Genomics 283(6):551–563
Kumar A, Hirochika H (2001) Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci 6(3):127–134
Lashermes P, Combes MC, Trouslot P, Charrier A (1997) Phylogenetic relationships of coffee-tree species (Coffea L.) as inferred from ITS sequences of nuclear ribosomal DNA. Theor Appl Genet 94(6–7):947–955
Leroy JF (1980) Evolution et taxogenèse chez les caféiers. Hypothèse sur leur origine. Comptes Rendus Hebdomadaires des Séances de l’Académie des Sciences. Série D 291(6):593–596
Leroy T, Marraccini P, Dufour M, Montagnon C, Lashermes P, Sabau X, Ferreira LP, Jourdan I, Pot D, Andrade AC, Glaszmann JC, Vieira LG, Piffanelli P (2005) Construction and characterization of a Coffea canephora BAC library to study the organization of sucrose biosynthesis genes. Theor Appl Genet 111(6):1032–1041
Louarn J (1992) La fertilite des hybrides interspécifiques et les relations génomiques entre caféiers diploïdesd’origine africaine (Genre Coffea L., sous-genre Coffea). Ph.D. Thesis, Paris XI Univ, p 200
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3(2):91–99
Mahesh V, Rakotomalala JJ, Le Gal L, Vigne H, de Kochko A, Hamon S, Noirot M, Campa C (2006) Isolation and genetic mapping of a Coffea canephora phenylalanine ammonia-lyase gene (CcPAL1) and its involvement in the accumulation of caffeoyl quinic acids. Plant Cell Rep 25(9):986–992
Maurin O, Davis AP, Chester M, Mvungi EF, Jaufeerally-Fakim Y, Fay MF (2007) Towards a phylogeny for Coffea (Rubiaceae): identifying well-supported lineages based on nuclear and plastid DNA sequences. Ann Bot (Lond) 100(7):1565–1583
Melayah D, Bonnivard E, Chalhoub B, Audeon C, Grandbastien MA (2001) The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J 28(2):159–168
Musoli P, Cubry P, Aluka P, Billot C, Dufour M, De Bellis F, Pot D, Bieysse D, Charrier A, Leroy T (2009) Genetic differentiation of wild and cultivated populations: diversity of Coffea canephora Pierre in Uganda. Genome 52(7):634–646
N’Diaye A, Poncet V, Louarn J, Hamon S, Noirot M (2005) Genetic differentiation between Coffea liberica var liberica and C. liberica var. Dewevrei and comparison with C. canephora. Plant Syst Evol 253:95–104
Natali L, Giordani T, Buti M, Cavallini A (2007) Isolation of Ty1-copia putative LTR sequences and their use as a tool to analyse genetic diversity in Olea europaea. Mol Breed 19(3):255–265
Noirot M, Poncet V, Barre P, Hamon P, Hamon S, de Kochko A (2003) Genome size variations in diploid African Coffea species. Ann Bot (Lond) 92(5):709–714
Pearce SR, Knox M, Ellis TH, Flavell AJ, Kumar A (2000) Pea Ty1-copia group retrotransposons: transpositional activity and use as markers to study genetic diversity in Pisum. Mol Gen Genet 263(6):898–907
Petit M, Lim KY, Julio E, Poncet C, Dorlhac de Borne F, Kovarik A, Leitch AR, Grandbastien MA, Mhiri C (2007) Differential impact of retrotransposon populations on the genome of allotetraploid tobacco (Nicotiana tabacum). Mol Genet Genomics 278(1):1–15
Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, Panaud O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16(10):1262–1269
Porceddu A, Albertini E, Barcaccia G, Marconi G, Bertoli FB, Veronesi F (2002) Development of S-SAP markers based on an LTR-like sequence from Medicago sativa L. Mol Genet Genomics 267(1):107–114
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277
Robbrecht E, Manen J-F (2006) The major evolutionary lineages of the coffee family (Rubiaceae, angiosperms). Combined analysis (nDNA and cpDNA) to infer the position of Coptosapelta and Luculia, and supertree construction based on rbcL, rps16, trnL-trnF and atpB-rbcL data. A new classification in two subfamilies, Cinchonoideae and Rubioideae. Syst Geogr Pl 76:85–146
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16(10):944–945
Sanz AM, Gonzalez SG, Syed NH, Suso MJ, Saldana CC, Flavell AJ (2007) Genetic diversity analysis in Vicia species using retrotransposon-based SSAP markers. Mol Genet Genomics 278(4):433–441
Schulman A (2007) Molecular markers to assess genetic diversity. Euphytica 158:313–321
Sonnhammer EL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167(1–2):GC1–GC10
Tam SM, Causse M, Garchery C, Burck H, Mhiri C, Grandbastien MA (2007) The distribution of copia-type retrotransposons and the evolutionary history of tomato and related wild species. J Evol Biol 20(3):1056–1072
Venturi S, Dondini L, Donini P, Sansavini S (2006) Retrotransposon characterisation and fingerprinting of apple clones by S-SAP markers. Theor Appl Genet 112(3):440–444
Vitte C, Panaud O (2005) LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet Genome Res 110(1–4):91–107
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BB, Powell W (1997) Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253(6):687–694
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8(12):973–982
Acknowledgments
The authors thank Mr. Francis Richard for maintaining the plants in the IRD greenhouses in Montpellier (France), and Dr. Pascal Musoli (NARO/COREC, Uganda) and Dr. Thierry Leroy (CIRAD, France) for kindly providing the Ugandan C. canephora DNAs.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M.-A. Grandbastien.
Rights and permissions
About this article
Cite this article
Hamon, P., Duroy, PO., Dubreuil-Tranchant, C. et al. Two novel Ty1-copia retrotransposons isolated from coffee trees can effectively reveal evolutionary relationships in the Coffea genus (Rubiaceae). Mol Genet Genomics 285, 447–460 (2011). https://doi.org/10.1007/s00438-011-0617-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-011-0617-0