Abstract
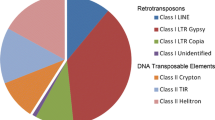
The (non-LTR) LINE and Ty3-gypsy-type LTR retrotransposon populations of three Vicia species that differ in genome size (Vicia faba, Vicia melanops and Vicia sativa) have been characterised. In each species the LINE retrotransposons comprise a complex, very heterogeneous set of sequences, while the Ty3-gypsy elements are much more homogeneous. Copy numbers of all three retrotransposon groups (Ty1-copia, Ty3-gypsy and LINE) in these species have been estimated by random genomic sequencing and Southern hybridisation analysis. The Ty3-gypsy elements are extremely numerous in all species, accounting for 18–35% of their genomes. The Ty1-copia group elements are somewhat less abundant and LINE elements are present in still lower amounts. Collectively, 20–45% of the genomes of these three Vicia species are comprised of retrotransposons. These data show that the three retrotransposon groups have proliferated to different extents in members of the Vicia genus and high proliferation has been associated with homogenisation of the retrotransposon population.





Similar content being viewed by others
References
Bennetzen JL, SanMiguel P, Chen M, Tikhonov A, Francki M, Avramova Z (1998) Grass genomes. Proc Natl Acad Sci USA 95:1975–1978
Chavanne F, Zhang DX, Liaud MF, Cerff R (1998) Structure and evolution of Cyclops: a novel giant retrotransposon of the Ty3/Gypsy family highly amplified in pea and other legume species. Plant Mol Biol 37:363–375
Flavell AJ, Smith DB, Kumar A (1992a) Extreme heterogeneity of Ty-copia group retrotransposons in plants. Mol Gen Genet 231:233–242
Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A (1992b). Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res 20:3639–3644
Gribbon BM, Pearce SR, Kalendar R, Schulman AH, Paulin L, Jack P, Kumar A, Flavell AJ (1999) Phylogeny and transpositional activity of Ty1-copia group retrotransposons in cereal genomes. Mol Gen Genet 261:883–891
Hattori M, et al (2000) The DNA sequence of human chromosome 21. Nature 405:311–319
Hill P (2003) Retrotransposons as determinants of genome size in three Vicia species. PhD Thesis, University of Dundee
Hirochika H, Hirochika R (1993) Ty1-copia group retrotransposons as ubiquitous components of plant genomes. Jpn J Genet 68:35–46
Kalendar R, Tanskanen J, Immolen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimate divergence. Proc Natl Acad Sci USA 97:6603–6607
Kato A, Iida Y, Yakura K, Tanifuji S (1985) Sequence analysis of Vicia faba highly repeated DNA: the BamHI repeated sequence families. Plant Mol Biol 5:41–53
Konieczny A, Voytas DF, Cummings MP, Ausubel FM (1991) A superfamily of Arabidopsis thaliana retrotransposons. Genetics 127:801–809
Kubis S, Heslop-Harrison JS, Desel C, Schmidt T (1998) The genomic organisation of non-LTR retrotransposons from three Beta species and five other Angiosperms. Plant Mol Biol 33:11–21
Kumar A. Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumekawa N, Ohtsubo E, Ohtsubo H (1999) Identification and phylogenetic analysis of gypsy-type retrotransposons in the plant kingdom. Genes Genet Syst 74:299–307
Laten HM, Majumdar A, Gaucher EA (1998) SIRE-1, a copia/Ty1-like retroelement form soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci USA 95:6897–6902
Lee D, Ellis THN, Turner L, Hellen RP, Cleary WG (1990) A Copia-like element in Pisum demonstrates the use of dispersed repeat sequences in genetic analysis. Plant Mol Biol 15:707–722
Manninen I, Schulman AH (1993) BARE-1 a copia-like retroelement in barley (Hordeum vulgare L.). Plant Mol Biol 22:829–846
Marracci S, Batistoni R, Pesole G, Citt L, Nardi I (1996) gypsy/Ty3-like elements in the genome of the terrestrial salamander Hydromantes (Amphibia, Urodela). J Mol Evol 43:584–593
Martin DMA., Hill P, Barton GJ, Flavell A.J (2003) Visual representation of database search results: the RHIMS plot. Bioinformatics 19:1–2
McCarthy EM, Liu J, Lizhi G, McDonald JF (2002) Long terminal repeat retrotransposons of Oryza sativa. Genome Biol Res 3:0053.1–0053.11
Neuman P, Pozarkova D, Macas J (2003) Highly abundant pea retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol Biol 53:399–410
Noma K, Ohtsubo E, Ohtsubo H (1999) Non-LTR retrotransposons (LINEs) as ubiquitous components of plant genomes. Mol Gen Genet 261:71–79
Nouzova M, Neuman P, Navratilova A, Galbraith DW, Macas J (2000) Microarray-based survey of repetitive genomic sequences in Vicia spp. Plant Mol Biol 45:229–244
Ohtsubo H, Kumekawa N, Ohtsubo E (1999) Rire2, a novel gypsy-type retrotransposon from rice. Genes Genet Syst 74:83–91
Pearce SR, Harrison G, Li D, Heslop-Harrison JS, Kumar A, Flavell AJ (1996) The Ty1-copia group retrotransposons in Vicia species: copy number, sequence heterogeneity and chromosomal localisation. Mol Gen Genet 250:305–315
Sambrook J, Fritsch EF Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
SanMiguel P, Gaut BS, Tikhonov A, Nakijama Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20:43–45
Sant VJ, Sainani MN, Sami-Subbu R, Ranjekar PK, Gupt VS (2000) Ty1-copia retrotransposon-like elements in chickpea genome: their identification, distribution and use for diversity analysis. Gene 257:157–166
Smit FA (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 657–663
Suoniemi A, Tanskanen J, Schulman AH. (1998) Gypsy retrotransposons are widespread in the plant kingdom. Plant J 13:699–705
Thompson JD Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vanderwiel PL, Voytas D, Wendel JF (1993) Copia-like retrotransposable element evolution in diploid and polyploid cotton (Gossypium L.). J Mol Evol 36:429–447
Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharov A, Schulman AH (1999) Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11:1769–1784
Wright DA, Voytas DF (1998) Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics 149:703–715
Wright DA, Ke N, Smalle J, Hauge BM, Goodman HM, Voytas DF (1996) Multiple non-LTR retrotransposons in the genome of Arabidopsis thaliana. Genetics 142:569–578
Acknowledgements
PH was funded by a BBSRC Earmark Studentship. This work was supported in part by Grants 960508 (TEBIODIV), 31502 (TEGERM) and FP6-2002-FOOD-1-506223 (GRAIN LEGUMES) from the European Commission under the Frameworks IV, V and VI. We thank Jeff Bennetzen for providing Ty3-gypsy group retrotransposon sequences prior to database submission, Steve Pearce for much helpful advice with the work, Gavin Ramsay for interesting discussions on Vicia phylogeny and for Vicia seeds, and Noel Ellis, Alan Schulman, Amar Kumar, Robbie Waugh and David Marshall for many interesting discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M.-A. Grandbastien
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Hill, P., Burford, D., Martin, D.M. et al. Retrotransposon populations of Vicia species with varying genome size. Mol Genet Genomics 273, 371–381 (2005). https://doi.org/10.1007/s00438-005-1141-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-1141-x