Abstract
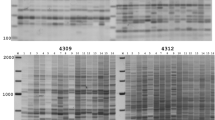
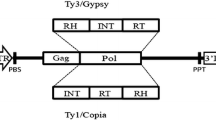
The BARE-1 retrotransposon is an active, dispersed, and highly abundant component of the genome of barley (Hordeum vulgare) and other species in its genus. Like all retrotransposons of its kind, BARE-1 is bounded by long terminal repeats (LTRs). We have developed two amplification-based marker methods based on the position of given LTRs within the genome. The IRAP (Inter-Retrotransposon Amplified Polymorphism) markers are generated by the proximity of two LTRs using outward-facing primers annealing to LTR target sequences. In REMAP (REtrotransposon-Microsatellite Amplified Polymorphism), amplification between LTRs proximal to simple sequence repeats such as constitute microsatellites produces markers. The methods can distinguish between barley varieties and produce fingerprint patterns for species across the genus. The patterns indicate that although the BARE-1 family of retrotransposons is disperse, these elements are locally clustered or nested and often found near tandem arrays of a simple sequence repeat.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 30 June 1998 / Accepted: 21 August 1998
Rights and permissions
About this article
Cite this article
Kalendar, R., Grob, T., Regina, M. et al. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98, 704–711 (1999). https://doi.org/10.1007/s001220051124
Issue Date:
DOI: https://doi.org/10.1007/s001220051124