Abstract
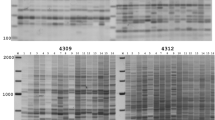
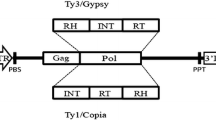
Retrotransposons are present in high copy number in many plant genomes. They show a considerable degree of sequence heterogeneity and insertional polymorphism, both within and between species. We describe here a polymerase chain reaction (PCR)-based method which exploits this polymorphism for the generation of molecular markers in barley. The method produces amplified fragments containing a Bare–1-like retrotransposon long terminal repeat (LTR) sequence at one end and a flanking host restriction site at the other. The level of polymorphism is higher than that revealed by amplified fragment length polymorphism (AFLP) in barley. Segregation data for 55 fragments, which were polymorphic in a doubled haploid barley population, were analysed alongside an existing framework of some 400 other markers. The markers showed a widespread distribution over the seven linkage groups, which is consistent with the distribution of the Bare–1 class of retrotransposons in the barley genome based on in situ hybridisation data. The potential applicability of this method to the mapping of other multicopy sequences in plants is discussed.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 17 July 1996 / Accepted: 20 September 1996
Rights and permissions
About this article
Cite this article
Waugh, R., McLean, K., Flavell, A. et al. Genetic distribution of Bare–1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253, 687–694 (1997). https://doi.org/10.1007/s004380050372
Issue Date:
DOI: https://doi.org/10.1007/s004380050372