Abstract
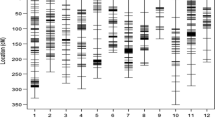
Diversity arrays technology (DArT) genomic libraries were developed from H. chilense accessions to support robust genotyping of this species and a novel crop comprising H. chilense genome (e.g., tritordeums). Over 11,000 DArT clones were obtained using two complexity reduction methods. A subset of 2,209 DArT markers was identified on the arrays containing these clones as polymorphic between parents and segregating in a population of 92 recombinant inbred lines (RIL) developed from the cross between H. chilense accessions H1 and H7. Using the segregation data a high-density map of 1,503 cM was constructed with average inter-bin density of 2.33 cM. A subset of DArT markers was also mapped physically using a set of wheat–H. chilense chromosome addition lines. It allowed the unambiguous assignment of linkage groups to chromosomes. Four segregation distortion regions (SDRs) were found on the chromosomes 2Hch, 3Hch and 5Hch in agreement with previous findings in barley. The new map improves the genome coverage of previous H. chilense maps. H. chilense-derived DArT markers will enable further genetic studies in ongoing projects on hybrid wheat, seed carotenoid content improvement or tritordeum breeding program. Besides, the genetic map reported here will be very useful as the basis to develop comparative genomics studies with barley and model species.



Similar content being viewed by others
References
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden M, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113:1409–1420
Alsop BP, Farre A, Wenzl P, Wang JM, Zhou MX, Romagosa I, Kilian A, Steffenson BJ (2011) Development of wild barley-derived DArT markers and their integration into a barley consensus map. Mol Breed 27:77–92
Alvarez JB, Martin A, Martin LM (1999a) Allelic variation of the d-prolamin subunits encoded at the Hch genome in a collection of primary hexaploid tritordeums. Theor Appl Genet 99:296–299
Alvarez JB, Martin LM, Martin A (1999b) Genetic variation for carotenoid pigment content in the amphiploid Hordeum chilense × Triticum turgidum conv. durum. Plant Breed 118:187–189
Alvarez JB, Caballero L, Nadal S, Ramírez MC, Martín A (2009) Development and gluten strength evaluation of introgression lines of Triticum urartu in durum wheat. Cereal Res Commun 37:243–248
Atienza SG, Gimenez MJ, Martin A, Martin LM (2000) Variability in monomeric prolamins in Hordeum chilense. Theor Appl Genet 101:970–976
Atienza SG, Ramirez CM, Hernandez P, Martin A (2004) Chromosomal location of genes for carotenoid pigments in Hordeum chilense. Plant Breed 123:303–304
Atienza SG, Satovic Z, Martin A, Martin LM (2005) Genetic diversity in Hordeum chilense Roem. et Schult. germplasm collection as determined by endosperm storage proteins. Genet Res Crop Evol 52:127–135
Atienza SG, Avila CM, Martin A (2007a) The development of a PCR-based marker for PSY1 from Hordeum chilense, a candidate gene for carotenoid content accumulation in tritordeum seeds. Aust J Agric Res 58:767–773
Atienza SG, Ballesteros J, Martin A, Hornero-Mendez D (2007b) Genetic variability of carotenoid concentration and degree of esterification among tritordeum (×Tritordeum Ascherson et Graebner) and durum wheat accessions. J Agric Food Chem 55:4244–4251
Atienza SG, Martin AC, Martin A (2007c) Introgression of wheat chromosome 2D or 5D into tritordeum leads to free-threshing habit. Genome 50:994–1000
Atienza SG, Martin AC, Ramirez MC, Martin A, Ballesteros J (2007d) Effects of Hordeum chilense cytoplasm on agronomic traits in common wheat. Plant Breed 126:5–8
Badea A, Eudes F, Salmon D, Tuvesson S, Vrolijk A, Larsson CT, Caig V, Huttner E, Kilian A, Laroche A (2011) Development and assessment of DArT markers in triticale. Theor Appl Genet 122:1547–1560
Ballesteros J, Ramirez MC, Martinez C, Atienza SG, Martin A (2005) Registration of HT621, a high carotenoid content tritordeum germplasm line. Crop Sci 45:2662–2663
Bartoš J, Sandve S, Kölliker R, Kopecký D, Christelová P, Stočes Š, Østrem L, Larsen A, Kilian A, Rognli O-A, Doležel J (2011) Genetic mapping of DArT markers in the Festuca/Lolium complex and their use in freezing tolerance association analysis. Theor Appl Genet 122:1133–1147
Bolibok-Bragoszewska H, Heller-Uszynska K, Wenzl P, Uszynski G, Kilian A, Rakoczy-Trojanowska M (2009) DArT markers for the rye genome—genetic diversity and mapping. BMC Genomics 10:578
Castillo A, Budak H, Varshney R, Dorado G, Graner A, Hernandez P (2008) Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol 8:97
Castillo A, Budak H, Martin AC, Dorado G, Borner A, Roder M, Hernandez P (2010a) Interspecies and intergenus transferability of barley and wheat D-genome microsatellite markers. Ann Appl Biol 156:347–356
Castillo A, Dorado G, Feuillet C, Sourdille P, Hernandez P (2010b) Genetic structure and ecogeographical adaptation in wild barley (Hordeum chilense Roemer et Schultes) as revealed by microsatellite markers. BMC Plant Biol 10:266
Cherif-Mouaki S, Said M, Alvarez JB, Cabrera A (2011) Sub-arm location of prolamin and EST-SSR loci on chromosome 1Hch from Hordeum chilense. Euphytica 178:63–69
Close T, Bhat P, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson J, Wanamaker S, Bozdag S, Roose M, Moscou M, Chao S, Varshney R, Szucs P, Sato K, Hayes P, Matthews D, Kleinhofs A, Muehlbauer G, DeYoung J, Marshall D, Madishetty K, Fenton R, Condamine P, Graner A, Waugh R (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10:582
Francki M, Walker E, Crawford A, Broughton S, Ohm H, Barclay I, Wilson R, McLean R (2009) Comparison of genetic and cytogenetic maps of hexaploid wheat (Triticum aestivum L.) using SSR and DArT markers. Mol Genet Genomics 281:181–191
Gil-Humanes J, Piston F, Martin A, Barro F (2009) Comparative genomic analysis and expression of the APETALA2-like genes from barley, wheat, and barley–wheat amphiploids. BMC Plant Biol 9:66
Graner A, Jahoor A, Schondelmaier J, Siedler H, Pillen K, Fischbeck G, Wenzel G, Herrmann RG (1991) Construction of an RFLP map of barley. Theor Appl Genet 83:250–256
Hagras AAA, Kishii M, Sato K, Tanaka H, Tsujimoto H (2005a) Extended application of barley EST markers for the analysis of alien chromosomes added to wheat genetic background. Breed Sci 55:335–341
Hagras AAA, Kishii M, Tanaka H, Sato K, Tsujimoto H (2005b) Genomic differentiation of Hordeum chilense from H. vulgare as revealed by repetitive and EST sequences. Genes Genet Syst 80:147–159
Harushima Y, Nakagahra M, Yano M, Sasaki T, Kurata N (2001) A genome-wide survey of reproductive barriers in an intraspecific hybrid. Genetics 159:883–892
Hernández P, Dorado G, Prieto Giménez MJ, Ramírez MC, Laurie DA, Snape JW, Martin A (2001) A core map of Hordeum chilense and comparisons with maps of barley (Hordeum vulgare) and wheat (Triticum aestivum). Theor Appl Genet 102:1259–1264
Inostroza L, del Pozo A, Matus I, Castillo D, Hayes P, Machado S, Corey A (2009) Association mapping of plant height, yield, and yield stability in recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Mol Breed 23:365–376
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acid Res 29:e25
Kumar S, Gill B, Faris J (2007) Identification and characterization of segregation distortion loci along chromosome 5B in tetraploid wheat. Mol Genet Genomics 278:187–196
Li HB, Kilian A, Zhou MX, Wenzl P, Huttner E, Mendham N, McIntyre L, Vaillancourt RE (2010) Construction of a high-density composite map and comparative mapping of segregation distortion regions in barley. Mol Genet Genomics 284:319–331
Mace E, Xia L, Jordan D, Halloran K, Parh D, Huttner E, Wenzl P, Kilian A (2008) DArT markers: diversity analyses and mapping in Sorghum bicolor. BMC Genomics 9:26
Mantovani P, Maccaferri M, Sanguineti M, Tuberosa R, Catizone I, Wenzl P, Thomson B, Carling J, Huttner E, DeAmbrogio E, Kilian A (2008) An integrated DArT-SSR linkage map of durum wheat. Mol Breed 22:629–648
Marcel T, Varshney R, Barbieri M, Jafary H, de Kock M, Graner A, Niks R (2007) A high-density consensus map of barley to compare the distribution of QTLs for partial resistance to Puccinia hordei and of defence gene homologues. Theor Appl Genet 114:487–500
Martin A, Martínez C, Rubiales D, Ballesteros J (1996) Tritordeum: triticale’s new brother cereal. In: Güedes-Pinto H, Darvey N, Carnide VP (eds) Triticale: today and tomorrow. Kluwer, Dordrecht, pp 57–72
Martin A, Martín LM, Cabrera A, Ramírez MC, Giménez MJ, Rubiales P, Hernández P, Ballesteros J (1998) The potential of Hordeum chilense in breeding Triticeae species. In: Jaradat AA (ed) Triticeae III. Science Publications, Enfield, pp 377–386
Martin AC, Atienza SG, Ramirez MC, Barro F, Martin A (2008) Male fertility restoration of wheat in Hordeum chilense cytoplasm is associated with 6H(ch)S chromosome addition. Aust J Agric Res 59:206–213
Martin AC, Atienza SG, Ramirez MC, Barro F, Martin A (2009) Chromosome engineering in wheat to restore male fertility in the msH1 CMS system. Mol Breed 24:397–408
Martín AC, Atienza SG, Ramírez M, Barro F, Martin A (2010) Molecular and cytological characterization of an extra acrocentric chromosome that restores male fertility of wheat in the msH1 CMS system. Theor Appl Genet 121:1093–1101
Matus I, Corey A, Filichkin T, Hayes PM, Vales MI, Kling J, Riera-Lizarazu O, Sato K, Powell W, Waugh R (2003) Development and characterization of recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Genome 46:1010–1023
Nasuda S, Kikkawa Y, Ashida T, Rafiqul Islam AKM, Sato K, Endo TR (2005) Chromosomal assignment and deletion mapping of barley EST markers. Genes Genet Syst 80:357–366
Oliver R, Jellen E, Ladizinsky G, Korol A, Kilian A, Beard J, Dumlupinar Z, Wisniewski-Morehead N, Svedin E, Coon M, Redman R, Maughan P, Obert D, Jackson E (2011) New Diversity Arrays Technology (DArT) markers for tetraploid oat (Avena magna Murphy et Terrell) provide the first complete oat linkage map and markers linked to domestication genes from hexaploid A. sativa L. Theor Appl Genet. doi:10.1007/s00122-011-1656-y (in press)
Piston F, Dorado G, Martin A, Barro F (2004) Cloning and characterization of a gamma-3 hordein mRNA (cDNA) from Hordeum chilense (Roem. et Schult.). Theor Appl Genet 108:1359–1365
Piston F, Martin A, Dorado G, Barro F (2005) Cloning and molecular characterization of B-hordeins from Hordeum chilense (Roem. et Schult.). Theor Appl Genet 111:551–560
Piston F, Dorado G, Martin A, Barro F (2006) Cloning of nine γ-gliadin mRNAs (cDNAs) from wheat and the molecular characterization of comparative transcript levels of γ-gliadin subclasses. J Cereal Sci 43:120–128
Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, Charmet G, Salse J (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genomics 9:473–484
Rodríguez-Suárez C, Giménez MJ, Atienza SG (2010) Progress and perspectives for carotenoid accumulation in selected Triticeae species. Crop Pasture Sci 61:743–751
Rodríguez-Suárez C, Atienza SG, Pistón F (2011a) Allelic variation, alternative splicing and expression analysis of Psy1 gene in Hordeum chilense Roem. et Schult. PLoS ONE 6:e19885
Rodríguez-Suárez C, Giménez MJ, Ramírez MC, Martín AC, Gutierrez N, Ávila CM, Martín A, Atienza SG (2011b) Exploitation of nuclear and cytoplasm variability in Hordeum chilense for wheat breeding. Plant Genet Res Charact Util 9:313–316. doi:10.1017/S1479262111000402
Rodríguez-Suárez C, Ramírez MC, Martín A, Atienza SG (2011c) Applicability of chromosome-specific SSR wheat markers for the introgression of Triticum urartu in durum wheat breeding programmes. Plant Genet Res Charact Util 9:439–444. doi:10.1017/S147926211100061X
Said M, Cabrera A (2009) A physical map of chromosome 4Hch from H. chilense containing SSR, STS and EST-SSR molecular markers. Euphytica 169:253–259
Sato K, Nankaku N, Takeda K (2009) A high-density transcript linkage map of barley derived from a single population. Heredity 103:110–117
Stein N, Prasad M, Scholz U, Thiel T, Zhang H, Wolf M, Kota R, Varshney R, Perovic D, Grosse I, Graner A (2007) A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor Appl Genet 114:823–839
Supriya A, Senthilvel S, Nepolean T, Eshwar K, Rajaram V, Shaw R, Hash C, Kilian A, Yadav R, Narasu M (2011) Development of a molecular linkage map of pearl millet integrating DArT and SSR markers. Theor Appl Genet 123:239–250
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, giovannoni JJ, Grandillo S, Martin GB, Messeguer R, Miller JC, Miller L, Paterson AH, Pineda O, Röder MS, Wing RA, Wu W, Yound ND (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
Tinker N, Kilian A, Wight C, Heller-Uszynska K, Wenzl P, Rines H, Bjornstad A, Howarth C, Jannink J-L, Anderson J, Rossnagel B, Stuthman D, Sorrells M, Jackson E, Tuvesson S, Kolb F, Olsson O, Federizzi L, Carson M, Ohm H, Molnar S, Scoles G, Eckstein P, Bonman JM, Ceplitis A, Langdon T (2009) New DArT markers for oat provide enhanced map coverage and global germplasm characterization. BMC Genomics 10:39
Van Ginkel M, Ogbonnaya F (2007) Novel genetic diversity from synthetic wheats in breeding cultivars for changing production conditions. Field Crops Res 104:86–94
Van Ooijen J (2006) Joinmap 4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
Van Os H, Stam P, Visser R, Van Eck H (2005) RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet 112:30–40
Varshney R, Marcel T, Ramsay L, Russell J, Röder M, Stein N, Waugh R, Langridge P, Niks R, Graner A (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet 114:1091–1103
Vaz Patto MC, Aardse A, Buntjer J, Rubiales D, Martin A, Niks RE (2001) Morphology and AFLP markers suggest three Hordeum chilense ecotypes that differ in avoidance to rust fungi. Can J Bot 79:204–213
Vaz Patto MC, Rubiales D, Martín A, Hernández P, Lindhout P, Niks RE, Stam P (2003) QTL mapping provides evidence for lack of association of the avoidance of leaf rust in Hordeum chilense with stomata density. Theor Appl Genet 106:1283–1292
Von Bothmer R, Giles BE, Jacobsen N (1986) Crosses and genome relationship in the Hordeum patagonicum group. Genetica 71:75–80
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Warburton M, Crossa J, Franco J, Kazi M, Trethowan R, Rajaram S, Pfeiffer W, Zhang P, Dreisigacker S, Ginkel M (2006) Bringing wild relatives back into the family: recovering genetic diversity in CIMMYT improved wheat germplasm. Euphytica 149:289–301
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A (2004) Diversity Arrays Technology (DArT) for whole-genome profiling of barley. PNAS 101:9915–9920
Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesná J, Cakir M, Poulsen D, Wang J, Raman R, Smith K, Muehlbauer G, Chalmers K, Kleinhofs A, Huttner E, Kilian A (2006) A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics 7:206
Xian-Liang S, Xue-Zhen S, Tian-Zhen Z (2006) Segregation distortion and its effect on genetic mapping in plants. Chin J Agric Biotech 3:163–169
Acknowledgments
This research was supported by grants (to S.G. Atienza) AGL2008-03720, 200840I137 and P09-AGR-4817 from the Spanish Ministry of Science and Innovation (MSI), CSIC, Junta de Andalucía and FEDER. We are grateful to E. León for her technical assistance. C. Rodríguez-Suárez acknowledges financial support from CSIC (JAE-Doc program). The authors thank Prof. Tsujimoto (Tottori University, Japan), for providing barley EST primers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2011_1741_MOESM2_ESM.xls
Supplementary File 2. Physical mapping of 287 DArT markers using H. chilense (H1)-wheat (Chinese Spring) chromosome and telosomic addition lines. (XLS 146 kb)
122_2011_1741_MOESM3_ESM.xls
Supplementary File 3. Features of loci of the linkage map. Excel spreadsheet containing a list of all mapped loci and their features. Data include locus position (in both Kosambi and Haldane), physical mapping when available and segregation in the RIL population. (XLS 2946 kb)
122_2011_1741_MOESM4_ESM.pdf
Supplementary Fig. 1. Genetic linkage map of the cross H1 × H7 containing 2,032 loci. Loci with physical mapping are indicated with a chromosome-code. The estimated centromere position is indicated as a green segment. For chromosomes 6Hch and 7Hch, where the centromere was positioned in a single map position, markers mapping to the centromere are shown in red color. (PDF 56 kb)
Rights and permissions
About this article
Cite this article
Rodríguez-Suárez, C., Giménez, M.J., Gutiérrez, N. et al. Development of wild barley (Hordeum chilense)-derived DArT markers and their use into genetic and physical mapping. Theor Appl Genet 124, 713–722 (2012). https://doi.org/10.1007/s00122-011-1741-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1741-2