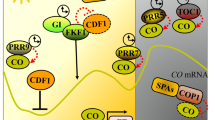
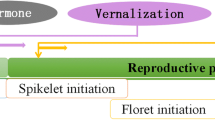
Abstract
The success of plant adaptation in different agroecological conditions is largely determined by the heading time. In grain crops, this is one of the basic traits in description of the variety, since the selection of varieties by the appropriate heading time for the region of their cultivation provides maximal realization of the potential of the variety by the productivity. To develop wheat varieties adapted to certain conditions, it is important to understand the mechanisms of formation of this trait at a diploid and polyploid level of genome organization. This review summarizes the main mechanisms underlying the formation of the heading time of diploid and polyploid wheat forms; the genes whose change in the expression is associated with variation in the common wheat heading time are described. Besides the main mechanisms such as photoperiod sensitivity and vernalization requirement, the effects of the circadian rhythms genes, phytohormones, light receptors, microRNA, and some other factors that significantly contribute to the heading time formation are analyzed.
Similar content being viewed by others
References
Putterill, J., Laurie, R., and Macknight, R., It’s time to flower: the genetic control of flowering time, Bioessays, 2004, vol. 26, pp. 363–373. doi 10.1002/bies.20021
Cockram, J., Jones, H., Leigh, F.J., et al., Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity, J. Exp. Bot., 2007, vol. 58, pp. 1231–1244. doi 10.1093/jxb/erm042
Blümel, M., Dally, N., and Jung, C., Flowering time regulation in crops–what did we learn from Arabidopsis?, Curr. Opin. Biotechnol., 2015, vol. 32, pp. 121–129. doi 10.1016/j.copbio.2014.11.023
Fowler, S., Lee, K., Onouchi, H., et al., GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains, EMBO J., 1999, vol. 18, pp. 4679–4688. doi 10.1093/emboj/18.17.4679
Mizoguchi, T., Wright, L., Fujiwara, S., et al., Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis, Plant Cell, 2005, vol. 17, pp. 2255–2270. doi 10.1105/tpc.105.033464
Simpson, G.G., The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time, Curr. Opin. Plant Biol., 2004, vol. 7, pp. 570–574. doi 10.1016/j.pbi.2004.07.002
He, Y., Chromatin regulation of flowering, Trends Plant Sci., 2012, vol. 17, pp. 556–562. doi 10.1016/j.tplants.2012.05.001
Kim, D.-H. and Sung, S., Genetic and epigenetic mechanisms underlying vernalization, Arabidopsis Book, 2014, vol. 12. e0171. doi 10.1199/tab.0171
Wahl, V., Ponnu, J., Schlereth, A., et al., Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana, Science, 2013, vol. 339, pp. 704–707. doi 10.1126/science.1230406
Aukerman, M.J. and Sakai, H., Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-Like target genes, Plant Cell, 2003, vol. 15, pp. 2730–2741. doi 10.1105/tpc.016238.pression
Chen, X., A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development, Science, 2004, vol. 303, pp. 2022–2025. doi 10.1126/science. 1088060
Jung, J.H., Seo, P.J., Ahn, J.H., et al., Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering, J. Biol. Chem., 2012, vol. 287, pp. 16007–16016. doi 10.1074/jbc.M111.337485
D’Aloia, M., Bonhomme, D., Bouché, F., et al., Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF, Plant J., 2011, vol. 65, pp. 972–979. doi 10.1111/j.1365-313X.2011.04482.x
Porri, A., Torti, S., Romera-Branchat, M., et al., Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods, Development, 2012, vol. 139, pp. 2198–2209. doi 10.1242/dev.077164
Johansson, M. and Staiger, D., Time to flower: interplay between photoperiod and the circadian clock, J. Exp. Bot., 2015, vol. 66, pp. 719–730. doi 10.1093/jxb/eru441
Song, Y.H., Shim, J.S., Kinmonth-Schultz, H.A., et al., Photoperiodic flowering: time measurement mechanisms in leaves, Annu. Rev. Plant Biol., 2015, vol. 66, pp. 441–464. doi 10.1146/annurev-arplant-043014-115555
Michaels, S.D. and Amasino, R.M., FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering, Plant Cell, 1999, vol. 11, pp. 949–956. doi 10.1105/tpc.11.5.949
Sheldon, C.C., Burn, J.E., Perez, P.P., et al., The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation, Plant Cell, 1999, vol. 11, pp. 445–458. doi 10.1105/tpc.11.3.445
Sheldon, C.C., Rouse, D.T., Finnegan, E.J., et al., The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC), Proc. Natl. Acad. Sci. U.S.A., 2000, vol. 97, pp. 3753–3758. doi 10.1073/pnas.060023597
Lee, J.H., Yoo, S.J., Park, S.H., et al., Role of SVP in the control of flowering time by ambient temperature in Arabidopsis, Genes Dev., 2007, vol. 21, pp. 397–402. doi 10.1101/gad.1518407
Posé, D., Verhage, L., Ott, F., et al., Temperaturedependent regulation of flowering by antagonistic FLM variant, Nature, 2013, vol. 503, pp. 414–417. doi 10.1038/nature12633
Thines, B.C., Youn, Y., Duarte, M.I., et al., The time of day effects of warm temperature on flowering time involve PIF4 and PIF5, J. Exp. Bot., 2014, vol. 65, pp. 1141–1151. doi 10.1093/jxb/ert487
Corbesier, L., Vincent, C., Jang, S., et al., FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis, Science, 2007, vol. 316, pp. 1030–1033. doi 10.1126/science.1141752
Yoo, S.K., Chung, K.S., Kim, J., et al., CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis, Plant Physiol., 2005, vol. 139, pp. 770–778. doi 10.1104/pp.105. 066928
Jaeger, K.E. and Wigge, P.A., FT protein acts as a long-range signal in Arabidopsis, Curr. Biol., 2007, vol. 17, pp. 1050–1054. doi 10.1016/j.cub.2007.05.008
Moon, J., Suh, S.S., Lee, H., et al., The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis, Plant J., 2003, vol. 35, pp. 613–623. doi 10.1046/j.1365-313X.2003.01833.x
Blázquez, M.A., Soowal, L.N., Lee, I., et al., LEAFY expression and flower initiation in Arabidopsis, Development, 1997, vol. 124, pp. 3835–3844.
Simon, R., Igeno, M.I., and Coupland, G., Activation of floral meristem identity genes in Arabidopsis, Nature, 1996, vol. 384, pp. 59–62. doi 10.1038/384059a0
Mandel, M.A. and Yanofsky, M.F., A gene triggering flower formation in Arabidopsis, Nature, 1995, vol. 377, pp. 522–524. doi 10.1038/377522a0
Ferrándiz, C., Gu, Q., Martienssen, R., et al., Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER, Development, 2000, vol. 127, pp. 725–734. doi 10.1046/j.1365-313x.1999.00442.x
Gu, Q., Ferrándiz, C., Yanofsky, M.F., et al., The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development, Development, 1998, vol. 125, pp. 1509–1517. doi 10.1105/tpc.1.1.37
Nitcher, R., Pearce, S., Tranquilli, G., et al., Effect of the Hope FT-B1 allele on wheat heading time and yield components, J. Hered., 2014, vol. 105, pp. 666–675. doi 10.1093/jhered/esu042
Würschum, T., Langer, S.M., and Longin, C.F.H., Genetic control of plant height in European winter wheat cultivars, Theor. Appl. Genet., 2015. doi 10.1007/s00122-015-2476-2
Worland, A.J., The influence of flowering time genes on environmental adaptability in European wheats vernalization sensitivity, Euphytica, 1996, vol. 89, pp. 49–57.
Pearce, S., Vanzetti, L.S., and Dubcovsky, J., Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1, Plant Physiol., 2013, vol. 163, pp. 1433–1445. doi 10.1104/pp.113.225854
Pearce, S., Kippes, N., Chen, A., et al., RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways, BMC Plant Biol., 2016, vol. 16, p. 141. doi 10.1186/s12870-016-0831-3
Chen, A., Li, C., Hu, W., et al., Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod, Proc. Natl. Acad. Sci. U.S.A., 2014, vol. 111, pp. 10037–10044. doi 10.1073/pnas.1409795111
Yan, L., Fu, D., Li, C., et al., The wheat and barley vernalization gene VRN3 is an orthologue of FT, Proc. Natl. Acad. Sci. U.S.A., 2006, vol. 103, pp. 19581–19586. doi 10.1073/pnas.0607142103
Chen, A. and Dubcovsky, J., Wheat TILLING mutants show that the vernalization gene VRN1 downregulates the flowering repressor VRN2 in leaves but is not essential for flowering, PLoS Genet., 2012, vol. 8. e1003134. doi 10.1371/journal.pgen.1003134
Galiba, G., Quarrie, S.A., Sutka, J., et al., RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat, Theor. Appl. Genet., 1995, vol. 90, pp. 1174–1179. doi 10.1007/BF00222940
Law, C.N., Worland, A.J., and Giorgi, B., The genetic control of ear-emergence time by chromosomes 5A and 5D of wheat, Heredity (Edinbourg), 1976, vol. 36, pp. 49–58. doi 10.1038/hdy.1976.5
Trevaskis, B., Bagnall, D.J., Ellis, M.H., et al., MADS box genes control vernalization-induced flowering in cereals, Proc. Natl. Acad. Sci. U.S.A., 2003, vol. 100, pp. 13099–13104. doi 10.1073/pnas.1635053100
Trevaskis, B., Hemming, M.N., Dennis, E.S., et al., The molecular basis of vernalization-induced flowering in cereals, Trends Plant Sci., 2007, vol. 12, pp. 352–357. doi 10.1016/j.tplants.2007.06.010
Kumar, S., Sharma, V., Chaudhary, S., et al., Genetics of flowering time in bread wheat Triticum aestivum: complementary interaction between vernalizationinsensitive and photoperiod-insensitive mutations imparts very early flowering habit to spring wheat, J. Genet., 2012, vol. 91, pp. 33–47.
Yan, L., Loukoianov, A., Tranquilli, G., et al., Positional cloning of the wheat vernalization gene VRN1, Proc. Natl. Acad. Sci. U.S.A., 2003, vol. 100, pp. 6263–6268. doi 10.1073/pnas.0937399100
Loukoianov, A., Yan, L., Blechl, A., et al., Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat, Plant Physiol., 2005, vol. 138, pp. 2364–2373. doi 10.1104/pp.105.064287
Greenup, A., Peacock, W.J., Dennis, E.S., et al., The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals, Ann. Bot., 2009, vol. 103, pp. 1165–1172. doi 10.1093/aob/mcp063
Yan, L., Helguera, M., Kato, K., et al., Allelic variation at the VRN-1 promoter region in polyploid wheat, Theor. Appl. Genet., 2004, vol. 109, pp. 1677–1186. doi 10.1007/s00122-004-1796-4
Fu, D., Szucs, P., Yan, L., et al., Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat, Mol. Genet. Genomics, 2005, vol. 273, pp. 54–65. doi 10.1007/s00438-004-1095-4
Shcherban, A.B., Efremova, T.T., and Salina, E.A., Identification of a new Vrn-B1 allele using two nearisogenic wheat lines with difference in heading time, Mol. Breed., 2012, vol. 29, pp. 675–685. doi 10.1007/s11032-011-9581-y
Potokina, E.K., Koshkin, V.A., Alekseeva, E.A., et al., The combination of the Ppd and Vrn gene alleles determines the heading date in common wheat varieties, Russ. J. Genet.: Appl. Res., 2012, vol. 2., no. 4, pp. 311–318.
Dubcovsky, J., Lijavetzky, D., Appendino, L., et al., Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement, Theor. Appl. Genet., 1998, vol. 97, pp. 968–975. doi 10.1007/s001220050978
Kippes, N., Chen, A., Zhang, X., et al., Development and characterization of a spring hexaploid wheat line with no functional VRN2 genes, Theor. Appl. Genet., 2016, vol. 129, pp. 1417–1428. doi 10.1007/s00122-016-2713-3
Hemming, M.N., Peacock, W.J., Dennis, E.S., et al., Integration of seasonal flowering time responses in temperate cereals, Plant Signal. Behav., 2008, vol. 3, pp. 601–602. doi 10.4161/psb.3.8.6352
Distelfeld, A., Li, C., and Dubcovsky, J., Regulation of flowering in temperate cereals, Curr. Opin. Plant Biol., 2009, vol. 12, pp. 178–184. doi 10.1016/j.pbi.2008.12.010
Li, C., Distelfeld, A., Comis, A., et al., Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes, Plant J., 2011, vol. 67, pp. 763–773. doi 10.1111/j.1365-313X.2011.04630.x
Evans, L.T., Short day induction of inflorescence initiation in some winter wheat varieties, Funct. Plant Biol., 1987, vol. 14, pp. 277–286.
Allard, V., Veisz, O., Kõszegi, B., et al., The quantitative response of wheat vernalization to environmental variables indicates that vernalization is not a response to cold temperature, J. Exp. Bot., 2012, vol. 63, pp. 847–857. doi 10.1093/jxb/err316
Dubcovsky, J., Loukoianov, A., Fu, D., et al., Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2, Plant Mol. Biol., 2006, vol. 60, pp. 469–480. doi 10.1007/s11103-005-4814-2
Pidal, B., Yan, L., Fu, D., et al., The CArG-box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response, J. Hered., 2009, vol. 100, pp. 355–364. doi 10.1093/jhered/esp002
Muterko, A., Kalendar, R., and Salina, E., Novel alleles of the VERNALIZATION1 genes in wheat are associated with modulation of DNA curvature and flexibility in the promoter region, BMC Plant Biol., 2016, vol. 16, p. 9. doi 10.1186/s12870-015-0691-2
Li, C. and Dubcovsky, J., Wheat FT protein regulates VRN1 transcription through interactions with FDL2, Plant J., 2008, vol. 55, pp. 543–554. doi 10.1111/j.1365-313X.2008.03526.x
Diallo, A.O., Ali-Benali, M.A., Badawi, M., et al., Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation, Mol. Genet. Genomics, 2012, vol. 287, pp. 575–590. doi 10.1007/s00438-012-0701-0
Golovnina, K.A., Kondratenko, E.Y., Blinov, A.G., et al., Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats, BMC Plant Biol., 2010, vol. 10, p. 168. doi 10.1186/1471-2229-10-168
Xiao, J., Xu, S., Li, C., et al., O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat, Nat. Commun., 2014, vol. 5, p. 4572. doi 10.1038/ncomms5572
Kippes, N., Debernardi, J.M., Vasquez-Gross, H.A., et al., Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia, Proc. Natl. Acad. Sci. U.S.A., 2015, vol. 112, pp. E5401–E5410. doi 10.1073/pnas.1514883112
Sung, S. and Amasino, R.M., Molecular genetic studies of the memory of winter, J. Exp. Bot., 2006, vol. 57, pp. 3369–3377. doi 10.1093/jxb/erl105
Pien, S. and Grossniklaus, U., Polycomb group and trithorax group proteins in Arabidopsis, Biochim. Biophys. Acta, 2007, vol. 1769, pp. 375–382. doi 10.1016/j.bbaexp.2007.01.010
Oliver, S.N., Finnegan, E.J., Dennis, E.S., et al., Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene, Proc. Natl. Acad. Sci. U.S.A., 2009, vol. 106, pp. 8386–8391. doi 10.1073/pnas. 0903566106
Yoshida, T., Nishida, H., Zhu, J., et al., Vrn-D4 is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat, Theor. Appl. Genet., 2010, vol. 120, pp. 543–552. doi 10.1007/s00122-009-1174-3
Thomas, B. and Vince-Prue, D., Photoperiodism in Plants, N.Y.: Acad. Press, 1996, 2nd ed.
Slafer, G.A. and Rawson, H.M., Sensitivity of wheat phasic development to major environmental factors: a reexamination of some assumptions made by physiologists and modellers, Aust. J. Plant Physiol., 1994, vol. 21, pp. 393–426. http://dx.doi.org/. doi 10.1071/PP9940393
Koshkin, V.A., Matvienko, I.I., Egorova, E.M., et al., The use of allele-specific markers of the Ppd-D1 gene for analysis of isogenic lines of spring common wheat, Tr. Prikl. Bot., Genet. Sel., 2009, vol. 166, pp. 151–156.
Beales, J., Turner, A., Griffiths, S., et al., A pseudoresponse regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.), Theor. Appl. Genet., 2007, vol. 115, pp. 721–733. doi 10.1007/s00122-007-0603-4
Borlaug, N.E., Contributions of conventional plant breeding to food production, Science, 1983, vol. 219, pp. 689–693. doi 10.1126/science.219.4585.689
Scarth, R. and Law, C.N., The location of the photoperiod gene, Ppd2 and an additional genetic factor for ear-emergence time on chromosome 2B of wheat, Heredity (Edinbourg), 1983, vol. 51, pp. 607–619. doi 10.1038/hdy.1983.73
Law, C.N., Sutka, J., and Worland, A.J., A genetic study of day-length response in wheat, Heredity (Edinbourg), 1978, vol. 41, pp. 185–191. doi 10.1038/hdy.1978.87
Wilhelm, E.P., Turner, A.S., and Laurie, D.A., Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.), Theor. Appl. Genet., 2009, vol. 118, pp. 285–294. doi 10.1007/s00122-008-0898-9
Worland, A.J., Börner, A., Korzun, V., et al., The influence of photoperiod genes on the adaptability of European winter wheats, Euphytica, 1998, vol. 100, pp. 385–394. doi 10.1023/A:1018327700985
Shaw, L.M., Turner, A.S., Herry, L., et al., Mutant alleles of Photoperiod-1 in wheat (Triticum aestivum L.) that confer a late flowering phenotype in long days, PLoS One, 2013, vol. 8. e79459. doi 10.1371/journal. pone.0079459
Worland, A.J., Borner, A., Korzun, V., et al., The influence of photoperiod genes on the adaptability of European winter wheats, Euphytica, 1998, vol. 100, pp. 385–394. doi 10.1023/a:1018327700985
Nishida, H., Yoshida, T., Kawakami, K., et al., Structural variation in the 5′ upstream region of photoperiod-insensitive alleles Ppd-A1a and Ppd-B1a identified in hexaploid wheat (Triticum aestivum L.), and their effect on heading time, Mol. Breed., 2013, vol. 31, pp. 27–37. doi 10.1007/s11032-012-9765-0
Díaz, A., Zikhali, M., Turner, A., et al., Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum), PLoS One, 2012, vol. 7. e33234. doi 10.1371/journal.pone.0033234
Kiss, T., Balla, K., Veisz, O., et al., Allele frequencies in the VRN-A1, VRN-B1 and VRN-D1 vernalization response and PPD-B1 and PPD-D1 photoperiod sensitivity genes, and their effects on heading in a diverse set of wheat cultivars (Triticum aestivum L.), Mol. Breed., 2014, vol. 34, pp. 297–310. doi 10.1007/s11032-014-0034-2
Shaw, L.M., Turner, A.S., and Laurie, D.A., The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum), Plant J., 2012, vol. 71, pp. 71–84. doi 10.1111/j.1365-313X.2012.04971.x
Suárez-López, P., Wheatley, K., Robson, F., et al., CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis, Nature, 2001, vol. 410, pp. 1116–1120.
Wu, L., Liu, D., Wu, J., et al., Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon, Plant Cell, 2013, vol. 25, pp. 4363–4377. doi 10.1105/tpc.113.118620
Peng, F.Y., Hu, Z., and Yang, R., Genome-wide comparative analysis of flowering-related genes in Arabidopsis, Wheat, and Barley, Int. J. Plant Genomics, 2015, vol. 2015, pp. 1–17. doi 10.1155/2015/874361
Takahashi, Y., Teshima, K.M., Yokoi, S., et al., Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice, Proc. Natl. Acad. Sci. U.S.A., 2009, vol. 106, pp. 4555–4560. doi 10.1073/pnas. 0812092106
Campoli, C., Drosse, B., Searle, I., et al., Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS, Plant J., 2012, vol. 69, pp. 868–880. doi 10.1111/j.1365-313X.2011.04839.x
Zhang, Z., Chen, J., Su, Y., et al., TaLHY, a 1R-MYB transcription factor, plays an important role in disease resistance against stripe rust fungus and ear heading in wheat, PLoS One, 2015, vol. 10, pp. 1–13. doi 10.1371/journal.pone.0127723
Strayer, C., Oyama, T., Schultz, T.F., et al., Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog, Science, 2000, vol. 289, pp. 768–771. doi 10.1126/science.289.5480.768
Alabadí, D., Oyama, T., Yanovsky, M.J., et al., Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock, Science, 2001, vol. 293, pp. 880–883. doi 10.1126/science.1061320
Zhao, X.Y., Hong, P., Wu, J.Y., et al., The taemiR408-mediated control of TaTOC1 genes transcription is required for the regulation of heading time in wheat, Plant Physiol., 2016, vol. 170, pp. 1578–1594. doi 10.1104/pp.15.01216
James, A.B., Monreal, J.A., Nimmo, G.A., et al., The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots, Science, 2008, vol. 322, pp. 1832–1835. doi 10.1126/science.1161403
Higgins, J.A., Bailey, P.C., and Laurie, D.A., Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses, PLoS One, 2010, vol. 5, p. 1. doi 10.1371/journal. pone.0010065
Zhang, W., Zhao, G., Gao, L., et al., Functional studies of heading date-related gene TaPRR73, a paralog of Ppd1 in common wheat, Front. Plant Sci., 2016, vol. 7, pp. 1–11. doi 10.3389/fpls.2016.00772
Calixto, C.P.G., Alternative splicing in the regulation of the barley circadian clock, Thesis PhD, Dundee: University of Dundee, 2013.
Campoli, C., Shtaya, M., Davis, S.J., et al., Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs, BMC Plant Biol., 2012, vol. 12, p. 97. doi 10.1186/1471-2229-12-97
Nusinow, D.A., Helfer, A., Hamilton, E.E., et al., The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth, Nature, 2011, vol. 475, pp. 398–402. doi 10.1038/nature10182
Gawronski, P., Ariyadasa, R., Himmelbach, A., et al., A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat, Genetics, 2014, vol. 196, pp. 1253–1261. doi 10.1534/genetics. 113.158444
Mizuno, N., Kinoshita, M., Kinoshita, S., et al., Loss-of-function mutations in three homoeologous PHYTOCLOCK 1 genes in common wheat are associated with the extra-early flowering phenotype, PLoS One, 2016, vol. 11. e0165618. doi 10.1371/journal. pone.0165618
Turner, A.S., Faure, S., Zhang, Y., et al., The effect of day-neutral mutations in barley and wheat on the interaction between photoperiod and vernalization, Theor. Appl. Genet., 2013, vol. 126, pp. 2267–2277. doi 10.1007/s00122-013-2133-6
Alvarez, M.A., Tranquilli, G., Lewis, S., et al., Genetic and physical mapping of the earliness per se locus Eps-Am1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene, Funct. Integr. Genomics, 2016, vol. 16, pp. 365–382. doi 10.1007/s10142-016-0490-3
Zikhali, M., Leverington-Waite, M., Fish, L., et al., Validation of a 1DL earliness per se (eps) flowering QTL in bread wheat (Triticum aestivum), Mol. Breed., 2014, vol. 34, pp. 1023–1033. doi 10.1007/s11032-014-0094-3
Wang, J., Wen, W., Hanif, M., et al., TaELF3-1DL, a homolog of ELF3, is associated with heading date in bread wheat, Mol. Breed., 2016, vol. 36, p. 161. doi 10.1007/s11032-016-0585-5
Zikhali, M., Wingen, L.U., and Griffiths, S., Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum), J. Exp. Bot., 2016, vol. 67, pp. 287–299. doi doi 10.1093/jxb/erv458
Yu, J.W., Rubio, V., Lee, N.Y., et al., COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability, Mol. Cell, 2008, vol. 32, pp. 617–630. doi 10.1016/j.molcel.2008.09.026
Xiang, Y.Z., Mao, S.L., Jia, R.L., et al., The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog, Plant Mol. Biol., 2005, vol. 58, pp. 53–64. doi 10.1007/s11103-005-4162-2
Sugano, S., Andronis, C., Green, R.M., et al., Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein, Proc. Natl. Acad. Sci. U.S.A., 1998, vol. 95, pp. 11020–11025. doi 10.1073/pnas.95.18.11020
Portolés, S. and Más, P., Altered oscillator function affects clock resonance and is responsible for the reduced day-length sensitivity of CKB4 overexpressing plants, Plant J., 2007, vol. 51, pp. 966–977. doi 10.1111/j.1365-313X.2007.03186.x
Ding, Z., Millar, A.J., Davis, A.M., et al., TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock, Plant Cell, 2007, vol. 19, pp. 1522–1536. doi 10.1105/tpc.106.047241
Para, A., Farré, E.M., Imaizumi, T., et al., PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock, Plant Cell, 2007, vol. 19, pp. 3462–3473. doi 10.1105/tpc.107.054775
Nakamichi, N., Kiba, T., Henriques, R., et al., PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock, Plant Cell, 2010, vol. 22, pp. 594–605. doi 10.1105/tpc.109.072892
Rawat, R., Takahashi, N., Hsu, P.Y., et al., REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock, PLoS Genet., 2011, vol. 7. e1001350. doi 10.1371/journal.pgen.1001350
Takase, T., Nishiyama, Y., Tanihigashi, H., et al., LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under noninductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1, Plant J., 2011, vol. 67, pp. 608–621. doi 10.1111/j.1365-313X.2011. 04618.x
Nelson, D.C., Lasswell, J., Rogg, L.E., et al., FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis, Cell, 2000, vol. 101, pp. 331–340. doi 10.1016/S0092-8674(00)80842-9
Somers, D.E., Kim, W., and Geng, R., The F-Box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time, Plant Cell, 2004, vol. 16, pp. 769–782. doi 10.1105/tpc.016808.1
Wu, J.-F., Wang, Y., and Wu, S.-H., Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering, Plant Physiol., 2008, vol. 148, pp. 948–959. doi 10.1104/pp.108.124917
Milec, Z., Valárik, M., Bartoš, J., et al., Can a late bloomer become an early bird? Tools for flowering time adjustment, Biotechnol. Adv., 2014, vol. 32, pp. 200–214. doi 10.1016/j.biotechadv.2013.09.008
Zanke, C., Ling, J., Plieske, J., et al., Genetic architecture of main effect QTL for heading date in European winter wheat, Front. Plant Sci., 2014, vol. 5, pp. 1–12. doi 10.3389/fpls.2014.00217
Bogard, M., Ravel, C., Paux, E., et al., Predictions of heading date in bread wheat (Triticum aestivum L.) using QTL-based parameters of an ecophysiological model, J. Exp. Bot., 2014. doi 10.1093/jxb/eru328
Perez-Lara, E., Semagn, K., Chen, H., et al., QTLs associated with agronomic traits in the Cutler × AC Barrie spring wheat mapping population using single nucleotide polymorphic markers, PLoS One, 2016, vol. 11. e0160623. doi 10.1371/journal.pone.0160623
Milner, S.G., Maccaferri, M., Huang, B.E., et al., A multiparental cross population for mapping QTL for agronomic traits in durum wheat (Triticum turgidum ssp. durum), Plant Biotechnol. J., 2015, vol. 14, pp. 1–14. doi 10.1111/pbi.12424
Cao, L., Hayashi, K., Tokui, M., et al., Detection of QTLs for traits associated with pre-harvest sprouting resistance in bread wheat (Triticum aestivum L.), Breed. Sci., 2016, vol. 66, pp. 260–270. doi 10.1270/jsbbs.66.260
Shirdelmoghanloo, H., Taylor, J.D., Lohraseb, I., et al., A QTL on the short arm of wheat (Triticum aestivum L.) chromosome 3B affects the stability of grain weight in plants exposed to a brief heat shock early in grain filling, BMC Plant Biol., 2016, vol. 16, p. 100. doi 10.1186/s12870-016-0784-6
Tahmasebi, S., Heidari, B., Pakniyat, H., et al., Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.), Genome, 2017, vol. 60, pp. 26–45. doi 10.1139/gen-2016-0017
Yu, M., Chen, G.-Y., Pu, Z.-E., et al., Quantitative trait locus mapping for growth duration and its timing components in wheat, Mol. Breed., 2015, vol. 35, p. 44. doi 10.1007/s11032-015-0201-0
Zou, J., Semagn, K., Iqbal, M., et al., Mapping QTLs controlling agronomic traits in the ‘Attila’ × ‘CDC Go’ spring wheat population under organic management using 90K SNP array, Crop Sci., 2017, vol. 57, p. 365. doi 10.2135/cropsci2016.06.0459
Zou, J., Semagn, K., Iqbal, M., et al., QTLs associated with agronomic traits in the Attila × CDC Go spring wheat population evaluated under conventional management, PLoS One, 2017, vol. 12. e0171528. doi 10.1371/journal.pone.0171528
Guo, Y., Du, Z., Chen, J., et al., QTL mapping of wheat plant architectural characteristics and their genetic relationship with seven QTLs conferring resistance to sheath blight, PLoS One, 2017, vol. 12. e0174939. doi 10.1371/journal.pone.0174939
Gerard, G.S., Börner, A., Lohwasser, U., et al., Genome-wide association mapping of genetic factors controlling Septoria tritici blotch resistance and their associations with plant height and heading date in wheat, Euphytica, 2017, vol. 213, p. 27. doi 10.1007/s10681-016-1820-1
Takahashi, Y., Shomura, A., Sasaki, T., et al., Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2, Proc. Natl. Acad. Sci. U.S.A., 2001, vol. 98, pp. 7922–7927. doi 10.1073/pnas.111136798
Kiseleva, A.A., Shcherban, A.B., Leonova, I.N., et al., Identification of new heading date determinants in wheat 5B chromosome, BMC Plant Biol., 2016, vol. 16(s1), pp. 35–46. doi 10.1186/s12870-015-0688-x
Suge, H. and Yamada, N., Flower-promoting effect of gibberellin in winter wheat and barley, Plant Cell Physiol., 1965, vol. 6, pp. 147–160.
Wilson, R.N., Heckman, J.W., and Somerville, C.R., Gibberellin is required for flowering in Arabidopsis thaliana under short days, Plant Physiol., 1992, vol. 100, pp. 403–408. doi 10.1104/pp.100.1.403
Li, A., Liu, D., Wu, J., et al., mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat, Plant Cell Online, 2014, vol. 26, pp. 1878–1900. doi 10.1105/tpc.114.124388
Kitagawa, S., Shimada, S., and Murai, K., Effect of Ppd-1 on the expression of flowering-time genes in vegetative and reproductive growth stages of wheat, Genes Genet. Syst., 2012, vol. 87, pp. 161–168.
Li, C., Lin, H., and Dubcovsky, J., Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley, Plant J., 2015, vol. 84, pp. 70–82. doi 10.1111/tpj.12960
Chen, F., Gao, M., Zhang, J., et al., Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China, BMC Plant Biol., 2013, vol. 13, p. 199. doi 10.1186/1471-2229-13-199
Lysenko, N.S., Kiseleva, A.A., Mitrofanova, O.P., et al., VIR World Collection Catalogue: Bread Wheat. Molecular Testing of the Vrn and Ppd Alleles in the Selection Varieties Approved for Use in the Russian Federation, St. Petersburg: VIR, 2014.
Zhang, X.K., Xiao, Y.G., Zhang, Y., et al., Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit, Crop Sci., 2008, vol. 48, pp. 458–470. doi 10.2135/cropsci2007.06.0355
Iqbal, M., Shahzad, A., and Ahmed, I., Allelic variation at the Vrn-A1, Vrn-B1, Vrn-D1, Vrn-B3 and Ppd-D1a loci of Pakistani spring wheat cultivars, Electron. J. Biotechnol., 2001, vol. 14, pp. 1–8. doi 10.2225/vol14-issue1-fulltext-6
Lv, B., Nitcher, R., Han, X., et al., Characterization of FLOWERING LOCUS T1 (FT1) gene in brachypodium and wheat, PLoS One, 2014, vol. 9. e94171. doi 10.1371/journal.pone.0094171
An, H., Roussot, C., Suárez-López, P., et al., CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis, Development, 2004, vol. 131, pp. 3615–3626. doi 10.1242/dev.01231
Wenkel, S., Turck, F., Singer, K., et al., CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis, Plant Cell, 2006, vol. 18, pp. 2971–2984. doi 10.1105/tpc.106.043299
Alqudah, A.M., Sharma, R., Pasam, R.K., et al., Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley, PLoS One, 2014, vol. 9. e113120. doi 10.1371/journal. pone.0113120
Yang, S., Murphy, R.L., Morishige, D.T., et al., Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12, PLoS One, 2014, vol. 9. e105352. doi 10.1371/journal. pone.0105352
Wang, J.W., Regulation of flowering time by the miR156-mediated age pathway, J. Exp. Bot., 2014, vol. 65, pp. 4723–4730. doi 10.1093/jxb/eru246
Takeda, T., Toyofuku, K., Matsukura, C., et al., Sugar transporters involved in flowering and grain development of rice, J. Plant Physiol., 2001, vol. 158, pp. 465–470. doi 10.1078/0176-1617-00358
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © A.A. Kiseleva, E.A. Salina, 2018, published in Genetika, 2018, Vol. 54, No. 4.
Rights and permissions
About this article
Cite this article
Kiseleva, A.A., Salina, E.A. Genetic Regulation of Common Wheat Heading Time. Russ J Genet 54, 375–388 (2018). https://doi.org/10.1134/S1022795418030067
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795418030067