Abstract
A proteomic study was conducted on caryopses of barley cv. Stratus (Hordeum vulgare). The caryopses were germinated in darkness at 20 °C, according to one of the three experimental setups: (a) in distilled water for 24 h, followed by 100 mM NaCl for another 24 h (salinity stress, SS); (b) in 100 µM abscisic acid for 24 h, with rinsing in distilled water to remove residual ABA, followed by 100 mM NaCl for another 24 h (pretreatment with ABA + salinity stress, ABAS); and (c) only in distilled water, for comparison with the tested samples (control, C). After 48 h, proteomes were extracted from barley sprouts in each treatment, according to the method by Gallardo et al. (2002a, b, modified). Proteins were then separated by 2DE and analyzed by MALDI–TOF/MS (Bruker). Under salinity stress, we observed changes in the pattern of proteins involved in: (a) energy metabolism, (b) one-carbon metabolism, (c) cell wall biogenesis, lignification, and adhesion, (d) response to oxidative stress, desiccation stress, and other defense responses, (e) structure and organization of the cytoskeleton, (f) signal transduction, transcription and translation processing, and (g) protein turnover. Our results revealed that ABA can alter the response of barley seedlings to salinity stress. In general, ABA pretreatment limited stress-induced proteomic changes. Alterations in energy and one-carbon metabolism indicate the importance of ABA in glycolytic/gluconeogenic pathways and the SMM cycle during salinity stress. Other observations demonstrated the role of ABA in hemicellulose, cellulose and lignin biosynthesis, ROS scavenging, actin CK formation, control of PI signaling, chromatin-mediated mechanism of stress tolerance, mRNA stability, and protection against dehydration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abscisic acid (ABA) is a hormone that is essential for the growth and development of plants. It is also a key mediator of physiological responses to environmental stressors. Plants have to cope with various abiotic stressors, including drought, cold, freezing, heat, and salinity. Crop productivity is considerably thwarted by salinity stress. An excess of mineral salts in soil could result from intensive bedrock weathering or scant precipitation, which prevents leaching of soluble mineral compounds from soil. In recent years, rapid climate changes, including higher temperatures and lower rainfalls, have exacerbated the problem of soil salinity in Europe. Unskillful fertilization can also cause transient soil salinity which is harmful to emerging plants. Soil salinity is induced by higher concentrations of potassium, magnesium and calcium salts, sulfuric acid, and salts of carbonic acid, which leads to excessive alkalization of the substrate and prevents iron uptake. However, soil salinity (sodicity) is largely determined by sodium chloride (NaCl), a soluble salt. Plants growing in saline environments are exposed to two main stressors: the osmotic effect (1) and salt ions which are accumulated in the cytoplasm (2). Under saline conditions, the osmotic potential of soil water decreases, which induces osmotic stress and a decrease in water uptake by plants. Therefore, salinity stress leads to dehydration of plant cells. This non-specific process is also observed in response to other abiotic stressors, such as drought, freezing, and mechanical damage. By contrast, the concentration of salt ions increases only in salinity stress. Moreover, salinity stress is always accompanied by non-specific, oxidative stress.
ABA is a plant stress hormone and a major endogenous messenger that induces high salinity responses and activates protective mechanisms against dehydration, toxic ions, and oxidation (Fahad et al. 2015). ABA is largely responsible for activating genes responsible for osmotic adjustment, ion compartmentalization and homeostasis, root hydraulic conductivity, membrane stability, shoot and root growth regulation, inhibition of transpiration rate, and wilting, thus contributing to the maintenance of the water and osmotic balance in plants (Lata and Prasad 2011). The discussed phytohormone regulates stomatal closure, thus minimizing transpiration and loss of water (Wilkinson and Davies 2010). Under abiotic stress, stomatal closure and gene expression which are induced by ABA involve Ca2+ and/or H2O2 as second messengers (Leung and Giraudat 1998; Chinnusamy et al. 2005; Zhang et al. 2007). The group of salt-responsive genes upregulated by ABA includes genes encoding: (1) dehydrins and late embryogenesis-abundant proteins (e.g., HVA1, RAB18, RD22, and AtADH), (2) vacuolar H+-inorganic pyrophosphatase (HVP1 and HVP10), tonoplast Na+/H+ antiporter (NHX1), the catalytic subunit (subunit A) of vacuolar H+-ATPase (HvVHA-A), and γ-tonoplast intrinsic protein-1 (TIP1), (3) mitogen-activated protein kinases (e.g., MAPK4 and OsMAPK5), germin-like protein-1 (GLP1), radical-scavenging antioxidant enzymes, including catalase (CAT), superoxide dismutase (SOD), glutathione reductase (GR), and ascorbate peroxidase (APX), and (4) enzymes that participate in the biosynthesis of osmoprotectants, such as proline and trehalose (Shi and Zhu 2002; Jiang and Zhang 2003; Chinnusamy et al. 2005; Fukuda and Tanaka 2006; Gomez et al. 2010; Keskin et al. 2010; Bakht et al. 2012; Sripinyowanich et al. 2013; Iqbal et al. 2014). Halophytes are plants that tolerate salt, whereas glycophytes such as wheat, rice, barley, soybeans, and potatoes are sensitive to saline conditions. Despite the above, barley is relatively resistant to saline environments (Mahmood 2011).
This study set out to determine the effect of salt stress on changes in the proteome of barley sprouts in an early growth stage. Abscisic acid (ABA) mediates the responses and/or adaptive changes to external stressors in the environment, therefore, the responses of ABA-pretreated barley sprouts to salinity stress were also monitored.
Materials and methods
Plant material and treatment conditions
The experiments were conducted on caryopses of barley cv. Stratus (Hordeum vulgare) supplied by the Plant Cultivation Station in Strzelce. Seeds were surface disinfected in 0.5% sodium hypochlorite for 20 min and washed with sterilized water. Twenty intact grains were placed on two layers of Whatman paper No. 1 (Whatman, Midstone, Kent, UK) in a Petri dish (ø 9 cm) and germinated in the dark at 20 °C under different conditions. Caryopses were germinated in distilled water for 24 h and immersed in 100 mM NaCl for another 24 h (salinity stress, SS). The remaining seeds were incubated in 100 μM abscisic acid for the first 24 h, rinsed with distilled water to remove residual ABA, and immersed in 100 mM NaCl for another day (ABA pretreatment + salinity stress, ABAS). Seeds germinated in distilled water for 48 h were the control (C). The examined material was composed of barley sprouts with a coleoptile emerged and a leaf at the tip. In agricultural practice, the above could be indicative of the germination phase, whereas the growth of a seedling starts with the emergence of the first leaf through the coleoptile and its unfolding. The phenotypes of the analyzed barley sprouts grown under different conditions are shown in Fig. 1.
Determination of sprout length, fresh and dry weight content
The length (coleoptiles, roots), fresh and dry weight of sprouts were determined in approximately 100 plants from at least three independent biological replicates (n = 3) of each sample (C, SS, and ABAS). Sprouts were separated from caryopses, surface drained on filter paper, measured, and weighed. To estimate dry weight content, sprouts were additionally dried at 85 °C for 48 h and weighed again. Root length was estimated for the longest root. The data were analyzed using Duncan’s test (Statistica 12) and expressed as mean ± SD. Differences in the length, fresh and dry weight of SS and C samples, and ABAS and SS samples were considered statistically significant at p ≤ 0.01.
Proteome extraction and 2D gel electrophoresis
Proteomes were extracted from the sprouts according to the method described by Gallardo et al. (2002a, b, modified). Young plants were isolated from 100 caryopses in each treatment and powdered in liquid nitrogen. The powdered tissue of each sample was suspended in chilled lysate buffer (10 µl per 1 mg of calculated DW) containing 7.5 M urea, 2.2 M thiourea, 63 mM CHAPS, 0.25% Triton X-100 (Bio-Rad), 5 mM Trizma base, 21 mM Trizma hydrochloride, 1.1% ampholytes (pH 3–10), 60 U/ml DNase I, 72 lg/ml RNase A (5.8 Kunitz units/ml), 14% protease inhibitors (Cocktail complete mini, Roche), and 8 mM DTT (Bio-Rad). Each sample was stirred, shaken on ice for 45 min and centrifuged several times at 12,000×g for 10 min to obtain clear supernatant. The protein extract was purified using the 2D Clean-Up Kit (GE Healthcare). Protein concentration was measured at 280 nm using a NanoDrop 1000 spectrophotometer (Thermo Scientific). 600 µg of the protein sample was solubilized in rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 0.5% ampholytes (pH 3–10) (Amersham), 10 mM DTT, and 0.002% bromophenol blue) to a total volume 450 µl. The mixture was loaded onto a 24 cm Immobiline DryStrip Gel strip with a linear pH gradient of 3–10 (GE Healthcare). IEF was performed in the Ettan IPGphor 3 system (GE Healthcare) according to the manufacturer’s instructions. Immediately prior to the second-dimensional run, the strip (containing separated proteins) was equilibrated in a glass tube in an equilibration buffer containing 6 M urea, 75 mM Tris–HCl (pH 6.8), 30% glycerol, and 26 mM DTT (in the first step) or 220 mM iodoacetamide (in the second step) for 15 min each. After equilibration, the strip was transferred to 12.5% SDS-PAGE gel (1 mm) and sealed with 10% agarose solubilized in an electrode buffer containing 25 mM Trizma base, 195 mM glycine, and 0.1% SDS (Laemmli 1970). Electrophoresis was carried in the EttanDalt Six electrophoresis unit (GE Healthcare) using the Laemmli buffer system, first at 2 W/gel for 0.5 h and then at 17 W/gel for the next 3 h. Low Molecular Mass Standards (Bio-Rad) were used to estimate the molecular masses of proteins.
Image and data analysis
After 2D-PAGE, the gel was stained with Coomassie Brilliant Blue G-250 according to Neuhoff et al. (1988) and scanned in an Image Scanner using the Labscan 3.0 software (GE Healthcare). Three images representing three independent biological replicates for each treatment (C, SS, and ABAS) were grouped and analyzed in the ImageMaster 2D Platinum 6.0 program. After spot detection, the quantitative determination of spot intensity, including data normalization before principal component analysis (Wibig 2007), was performed based on the average values of background optical density (OD) and spot intensity on each gel. The spot volumes in SS were compared with those in C, and the spot volumes in ABAS were compared with those in SS. Proteins exhibiting at least 1.5-fold reproducible changes in abundance between the compared samples were subjected to statistical analysis using Duncan’s parametric test (p ≤ 0.01). Proteins whose abundance changed significantly were excised from the gels manually and identified by mass spectrometry.
Protein identification
The spots which were cut out from the gels were washed and destained with 25 mM ammonium bicarbonate in 50% ethanol for 1 h at 37 °C. Before in-gel digestion, proteins were reduced with 10 mM DTT in 25 mM ammonium bicarbonate for 30′ at 60 °C, and alkylated with 10 mM iodoacetamide in 25 mM ammonium bicarbonate for 45′ at 37 °C in the dark. Proteins were dried in a vacuum centrifuge and digested using sequencing grade trypsin solution (Promega, Madison, USA) containing 10 ng/µl trypsin in 25 mM ammonium bicarbonate. Digestion was carried out at 37 °C overnight. After in-gel digestion, gels were washed three times with 0.1% TFA in 50% acetonitrile to collect protein peptides. After the extraction of peptide fragments from the gel, samples were desalted and cleaned with Zip-Tip C18 (Millipore) according to the manufacturer’s instructions. Tryptic mixed peptides in the amount of 1 µl were loaded onto an AnchorChip (Bruker) target plate, one volume of saturated CHCA (Sigma) in 50% v/v acetonitrile with 0.1% TFA was added, and the peptides were cocrystallized. The loaded target was analyzed in the MALDI–TOF/MS system (Bruker) equipped with a nitrogen laser and operated in a reflector/delayed extraction mode for MALDI–TOF PMF with the FlexControl software (Bruker). The instrument was calibrated externally with (M + H)+ ions of Bradykinin 1, Angiotensin II, Angiotensin I, Substance P, Bombesin, Renin Substrate, ACTH clip 1, ACTH clip 18, and Somatostatin 28 (Peptide Calibration Standard II, Bruker). The MS spectra were searched against the National Center for Biotechnology Information database (NCBI). The MASCOT search engine (http://www.matrixscience.com) was used for PMF search. Database queries were carried out using the following parameters: trypsin cleavage, taxonomy of “Viridiplantae (Green Plants)”, monoisotopic mass accuracy with 80 ppm tolerance, one missed cleavage site, carbamidomethylation of cysteine as fixed modification, and oxidation of methionine as variable modification. A protein was positively identified based on the following criteria: protein scores higher than 75 and minimum 10% protein coverage by the matching peptides, including at least three independent peptides with mass deviation of less than 50 ppm. Theoretical pI and MW values were calculated from the identified sequence in NCBInr using the ExPASy tool (http://www.expasy.org).
Results
Changes in the length, fresh and dry weight of barley sprouts
The effect of salinity on barley growth and the changes induced by ABA pretreatment were investigated based on differences in the length of coleoptiles and roots, as well as fresh and dry weight of sprouts. The investigated parameters were altered by the applied treatments, both in SS and in ABAS probes (Fig. 2a, b). In SS probes, the length, fresh and dry weight of stressed plants were reduced by around 45% (both coleoptile and root), 51 and 33%, respectively. These results indicate that the growth, dry weight accumulation, and hydration of seedling tissue were significantly inhibited under salinity stress. All of the examined parameters were low in SS, but the stressed sprouts had a characteristic morphology. The roots of stressed seedlings had more root hairs than control roots (Fig. 1). The inhibition of seedling growth under ABA pretreatment was manifested by primary root growth (by around 48%). Despite the above, changes in the length of coleoptiles or dry weight were not observed in the ABAS sample. However, the average fresh weight of sprouts in ABAS was approximately 28% higher than in SS.
Changes in the proteome under different treatments
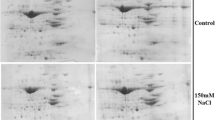
Our results revealed proteomic changes in barley seedlings treated with NaCl, indicating that ABA pretreatment altered the salinity response. To separate and identify differentially expressed proteins, IEF/SDS-PAGE coupled with MALDI TOF mass spectrometry was performed. In the group of matched spots, the abundance levels (% vol) of 52 protein spots were changed under saline conditions. However, 44 of these spots were successfully identified by spectrometric analysis (Fig. 3; Table 1). Based on their function, the identified salt-responsive proteins were classified as proteins involved in: (a) energy metabolism, (b) one-carbon metabolism, (c) cell wall biogenesis, lignification, and adhesion, (d) response to oxidative stress, desiccation stress, and other defense responses, (e) structure and organization of the cytoskeleton, (f) signal transduction, transcription, and translation processing, and (g) protein turnover (Table 1; Fig. 4).
2DE profiles of proteins from barley sprouts grown under control conditions (C), salt stress conditions (SS), and pretreated with ABA before exposure to salt stress (ABAS). Proteins were separated from each sample in the amount of 600 µg by 2DE and visualized by CBB G-250 staining. They were separated in pH 3–10 (24 cm, linear) in the first dimension and 12.5% SDS-PAGE in the second dimension. Significantly altered protein spots (p ≤ 0.01), whose volume% changed more than 1.5-fold, are marked with arrows (unchanged spots in ABAS are marked with dotted arrows). Altered spots were analyzed by MALDI–TOF/MS. The list of the identified proteins is shown in Table 1
In the group of 44 proteins regulated by salinity, 24 proteins were altered by ABA pretreatment. Exogenously applied ABA minimizes salinity-induced changes. Moreover, the expression of five proteins (spots 13, 14, 15, 26, and 35) in the ABAS probe was silenced to the extent comparable to that noted in the control treatment (Fig. 3; Table 1). The highest percentage of proteins altered by ABA was observed in the group of proteins involved in: (1) biogenesis and lignification of the cell wall and (2) responses to oxidative and desiccation stress. By contrast, ABA did not influence salt-responsive proteins classified as proteins that are involved in other defense responses and protein turnover (Fig. 4).
Discussion
High salt concentrations inhibit plant growth, development, and survival. Exposure to high salt concentrations influences major life processes in plants (Batool et al. 2014). Our research demonstrated that salinity stress significantly inhibited the growth of barley sprouts. However, the characteristic morphology of roots (with a higher number of root hairs) in stressed plants points to the initiation of protective processes against salinity. The aim of our work was to estimate processes that play a key role in salinity response, including the role of ABA in salt stress prevention. Our proteomic study revealed salinity-induced changes which can explain the morphological adaptation of sprouts to saline conditions. Incubation in 100 μM ABA which inhibits plant growth and development, followed by salinity stress, led to even greater inhibition of root growth, but not the growth or density of root hairs. However, the fresh weight of stressed sprouts preincubated in ABA increased. Jones et al. (1987) demonstrated that the growth of wheat roots was inhibited at ABA concentrations that increased turgor pressure. The above findings suggest that the physiological effect of salt stress was alleviated by ABA pretreatment, mainly by the regulation of the water balance in analyzed barley sprouts. In our study, ABA pretreatment limited stress-induced changes in the barley proteome. Our independent research (data not shown) demonstrated that the treatment of barley caryopses with 100 μM ABA induced proteomic changes similar to those caused by salinity, excluding the JIP protein. The observed reduction in the salt-responsive proteome could indicate that ABA pretreatment plays a role in the acquisition of tolerance to salinity stress. The metabolic functions of proteins affected by both salt and ABA are discussed below.
Energy metabolism
Under NaCl stress, plants decrease their metabolic rates to conserve energy and limit ROS production (Möller 2001; Golldack et al. 2014). Two enzymes involved in energy metabolism were downregulated: putative cytochrome c oxidase subunit II PS17 (spot 4) and ATP synthase beta subunit (spot 10). Cytochrome c oxidase is a terminal enzyme in the mitochondrial respiratory chain, whereas the ATP synthase beta subunit is a mitochondrial proton-transporting ATP synthase complex. The downregulation of these proteins could result from the disruptive influence of salinity stress on membranes.
However, not all proteins involved in catabolism were downregulated. For example, we observed the upregulation of the cytosolic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (spot 25) which plays a major role in glycolytic and gluconeogenic pathways. GAPDH catalyzes the reversible oxidation and phosphorylation of glyceraldehyde 3-phosphate into 3-phospho-d-glyceroyl phosphate. This enzyme is essential for the maintenance of cellular ATP levels and carbohydrate metabolism. GAPDH is synthesized in roots during anoxic conditions, and it belongs to the group of anaerobic polypeptides (Manjunath and Sachs 1997). It appears that GAPDH responds to NAD:NADH and ATP:ADP ratios in the cell to accommodate the cell’s specific requirements (Velasco et al. 1994). In addition to its catalytic role, GAPDH is also involved in responses to various abiotic stressors in plants (Yang Zhai 2017). Under stress, GAPDH plays a pivotal role in redox signaling by interacting with F-actin. Cytosolic GAPDH also interacts with the plasma membrane-associated phospholipase D (PLDδ) in H2O2-mediated signal transduction. Genetic ablation of PLDδ impeded stomatal response to abscisic acid and hydrogen peroxide, placing PLDδ downstream of H2O2 in mediating ABA-induced stomatal closure (Guo et al. 2012). In our study, GAPDH was the only energy metabolism enzyme to be affected by ABA treatment.
One-carbon metabolism
Methionine (Met) is metabolized in two ways: by (1) the activated methyl cycle or (2) the S-methylmethionine (SMM) cycle in one-carbon (C1) metabolism. In the first metabolic process, Met is transformed to S-adenosylmethionine (SAM) which participates in the synthesis of selected metabolites, such as glycine betaine, methylated polyols, and polyamines. Glycine betaine and polyols are osmoprotectants which are accumulated in response to water/salinity stress (Bohnert and Jensen 1996; Fujiwara et al. 2010). Polyamines participate in various processes at the cellular level, including DNA replication, transcription, translation, cell proliferation, modulation of enzymatic activity, membrane stability, cation–anion balance in cells, and wall lignification (Gill and Tuteja 2010; Ogawa and Mitsuya 2012).
In the second metabolic process, Met is converted to S-methylmethionine in the SMM cycle. SMM is synthesized when a methyl group is transferred from S-adenosylmethionine (SAM) to methionine (Met) in a reaction mediated by methionine methyltransferase (MMT), and it is reconverted to Met with homocysteine (Hcy) in a reaction catalyzed by homocysteine methyltransferase (HMT). Ranocha et al. (2001) suggested that the SMM cycle uses half of the synthesized SAM (1) to inhibit the accumulation of SAM. However, SMM is converted to 3-dimethylsulfoniopropionate (DMSP), a compatible solute in selected salt-tolerant plants (Hanson et al. 1994).
In our work, salinity induced the downregulation of two enzymes involved in the activated methyl cycle—methionine synthase (spot 5) and S-adenosylmethionine synthase (spot 24), and the upregulation of homocysteine S-methyltransferase (spot 28) involved in MMT. In this group of enzymes, only HMT was altered by ABA. This observation revealed changes in one-carbon metabolism, and it emphasized the importance of SMM in salinity response and ABA action. Our findings differ from the results reported in other studies, where the activated methyl cycle played the main role in responses to ABA and salt treatment. Espartero et al. (1994) suggested that SAM genes are responsive to salt stress and that ABA could participate in the upregulation of SAM transcripts.
Serine hydroxymethyltransferase (SHMT), an enzyme that catalyzes the reversible conversion of serine and tetrahydrofolate (THF) to glycine and N 5,N 10-methylene-THF, is also involved in the pathway for the interconversion of C-1 compounds. We identified SHMT (spot 19) as a salinity-upregulated protein which was also altered by ABA treatment. We also observed the upregulation of dehydration-responsive protein (spot 1), a predicted protein with S-adenosylmethionine-dependent activity. The increase in both proteins in the analyzed tissue emphasizes the significance of methylation processes during salinity stress. Less is known about the direct influence of ABA on SHMT, but Verslues et al. (2007) and Kaur et al. (2009) suggested links between sensitivity to ABA and glycolate metabolism, in particular the activity of peroxisomal glu:glyoxylate aminotransferase (GGT). GGT converts glycolate to glycine, the substrate for SHMT. GGT is essential for ABA-induced accumulation of proline and gene expression, and accumulation of ABA under exposure to stress and pretreatment with ABA. The results of this study validate the assumption that hydrogen peroxide participates in ABA signaling in proline accumulation.
Cell wall biogenesis, lignification, and adhesion
The cell walls of monocots, including wheat and barley, are composed of a cellulosic network embedded in a matrix of hemicellulose fibers and pectin polysaccharides. The major polysaccharides include glucuronoarabinoxylans and (1,3;1,4)-β-d-glucans, whereas the levels of glucomannans, xyloglucans, and pectic polysaccharides are relatively low (Farrokhi et al. 2006).
In our study, selected enzymes responsible for wall synthesis were upregulated under salinity stress. We identified two enzymes involved in hemicellulose synthesis: UDP-d-glucose dehydrogenase (spot 21) and xylose isomerase (spot 13). UDP-d-glucose dehydrogenase (UDPGDH) oxidizes UDP-d-glucose (UDP-Glc) to UDP-d-glucuronate (UDP-GlcA), the precursor of UDP-d-xylose and UDP-l-arabinose, and the main cell wall polysaccharide precursors. Xylose isomerase catalyzes the synthesis of d-xylose from d-xylulose, a precursor of xyloglucan, and hemicellulose.
Another detected enzyme, glucan endo-1-3-β-glucosidase GIII (spot 31), could play a role in the synthesis of cellulose. Several authors have demonstrated that hydrolytic enzymes are involved in the reorganization of newly synthesized cellulose chains (Szyjanowicz et al. 2004; Farrokhi et al. 2006). Moreover, the key quantitative trait locus (QTL) associated with the content of (1,3;1,4)-β-d-glucan in barley grain co-segregates with the gene encoding (1,3;1,4)-β-d-glucanase (Han et al. 1995).
We also identified pectin methyltransferase, probable methyltransferase PMT10 (spot 2) which could control cell wall adhesion by incorporating methyl groups into pectin in their synthesis site (Vannier et al. 1992; Goubet and Mohnen 1999). Pectic polysaccharides such as homogalacturonan (HGA) are produced in the Golgi apparatus. HGA comprises a linear chain of α-1,4-linked d-galacturonic acid (GalA) that undergoes different methyl esterification reactions at C6 (Bacic et al. 1988). The degree of pectin methyl esterification is a key determinant of pectin’s adhesive properties (Krupková et al. 2007).
We also observed the upregulation of three enzymes involved in lignin biosynthesis: putative anthocyanidin-3-glucoside rhamnosyltransferase (spot 12) and two peroxidases—peroxidase BP1 (spot 27) and peroxidase 2 (spot 37).
In the walls of grass cells, lignin is a polymer composed of phenylpropanoid monolignol units: hydroxycinnamic, ferulic, and p-coumaric acids. Ferulic acid and, partly, p-coumaric acid are esterified to arabinose molecules of arabinoxylans. Arabinoxylan polymers are also cross-linked through diferulates (Jung and Deetz 1993).
Oxidative stress caused by pathogenic infections, wounding, and other environmental stressors leads to oxidative burst and the release of ROS. Peroxidases catalyze the reduction of H2O2 by relying on cell wall molecules as substrates such as lignin precursors, phenolic compounds, or auxin, which leads to polymerization reactions, including suberization, lignification, and cell wall protein cross-linking (Kristensen et al. 1999; Passardi et al. 2004a, b). An increase in peroxidase levels could suggest that the cell wall is undergoing modification to reduce the influx of ions under saline conditions. Moreover, lignification is intensified by putative anthocyanidin-3-glucoside rhamnosyltransferase which is involved in the biosynthesis of flavonoids, stilbene and lignin. Peroxidases play an important role in the metabolism of apoplastic ROS, and they contribute to root growth and development. Hydrogen peroxide appears to be involved in root growth restriction and root hair formation (Dunand et al. 2007). In their oxygenated form (oxyperoxidases), peroxidases can increase H2O2 levels in the zone of differentiation in roots. The catalytic versatility of peroxidases can explain the morphological changes observed in barley roots in our study. Hydrogen peroxide could inhibit root growth and stimulate root hair formation under salt stress. The observed significant inhibition of root growth, accompanied by rapid formation of root hairs, indicates that ABA pretreatment enhances the mechanisms of resistance to environmental stress.
Our results suggest that the maintenance of cell wall architecture is crucial under stress and that ABA plays an important role in this process. The only protein that was not affected by abscisic acid was probable methyltransferase PMT10 (spot 2). These findings emphasize the importance of ABA in cellulose and hemicellulose synthesis and in lignification processes which increase the thickness of cell walls. Park et al. (2011) suggested a possible connection between the involvement of MYB52 genes in ABA response and cell wall biosynthesis. Other authors noted that ABA inhibits seed germination by preventing the loosening and expansion of cell walls (Gimeno-Gilles et al. 2009). Our results suggest that ABA induces cell wall thickening by increasing the biosynthesis of cell wall components, which could restrict elongation and enhance protective mechanisms against stress.
Response to oxidative stress
Other enzymes that catalyze ROS scavenging, including catalase (spot 11, 20) and glutathione peroxidase (spot 14), were also identified in this study. Roxas et al. (2000) reported that overexpression of glutathione S-transferase with glutathione peroxidase activity (GST/GPX) in transgenic plants promoted the growth of seedlings under exposure to various stressors. They demonstrated that seedlings expressing GST/GPX (GST+) were characterized by a high growth rate under exposure to low temperatures and saline stress, whereas the growth of control seedlings was stunted under the same conditions. In other studies, transgenic GST+ seedlings were significantly more abundant in oxidized glutathione (GSSG) under exposure to stress in comparison with wild-type seedlings (Roxas et al. 1997). High levels of GSSG could point to intensified scavenging activity of GPX-dependent peroxide in seedlings. The above could prevent cellular damage, promote metabolic activity, and stimulate growth in seedlings (Roxas et al. 2000). According to Miao et al. (2006), GPX3 plays a dual role: first, as a scavenger of H2O2, and second, as a transducer in ABA and H2O2 signaling pathways, where it regulates stomatal movement during drought.
In plant cells, the activity of ABA could be related to oxidative stress. This phytohormone could stimulate the production of H2O2 (Zhang et al. 2001; Jiang and Zhang 2001, 2003) and induce the expression of selected antioxidant genes encoding, for example, catalase (CAT) and peroxidase (POX) (Guan et al. 2000; Jiang and Zhang 2003; Agarwal et al. 2005). Low doses of ABA (10–100 μM) triggered an antioxidant defense mechanism to prevent oxidative damage to membrane lipids and proteins (Jiang and Zhang 2001). A minor, ABA-induced increase in H2O2 levels could be required to build stress tolerance and enhance survival in plants exposed to environmental stressors.
Structure and organization of the cytoskeleton
We identified two cytoskeletal proteins: tubulin alpha-1 chain (spot 23) and actin-58 (spot 26), as well as proteins that affect cytoskeleton (CK) organization: profilin1 (spot 40) and putative formin-like protein 15b (AtFH15b) (spot 36). All proteins were upregulated under saline conditions. Tubulin is a building block of the cytoskeleton, and it is involved in the intracellular transport of organelles, including chromosomes and mitotic spindles. The first group of compounds that are targeted by oxidative stress includes tubulin (Apraiz et al. 2006; Wu et al. 2013). In animal cells, β-tubulin can promote phosphorylation during microtubule formation, and it protects the cytoskeleton against oxidative stress (Howard and Hyman 2003). The cytoskeleton is also composed of microfilaments (MFs) which could play a role during salt stress. According to Zhao et al. (2013), stress signaling relies on the interactions between microfilaments and Ca2+ in plants. In bacteria, the actin cytoskeleton controls immune cell signaling and the production of cytokines and reactive O2− (Aktories et al. 2011).
Profilin is a ubiquitous protein which binds to actin and modifies cytoskeletal structure by polymerizing or depolymerizing actin filaments. It also participates in plant cell responses to internal and external signals (Fatehi et al. 2012). Profilin is involved in signaling pathways, which promotes rapid formation of actin filaments (Ridley et al. 2003; Lindberg et al. 2008; Yu 2008). Actin polymerization also involves formins which belong to a different family of multimodular proteins (Carlier and Pantaloni 2007). Formins associate with the fast-growing end of actin filaments and incorporate actin from profilin:actin structures (Romero et al. 2007; Sarmiento et al. 2008; Yu 2008).
In the present study, ABA affected two cytoskeletal proteins: actin and putative formin-like protein, which suggests that ABA influences the organization of actin filaments. The cytoskeleton is involved in signaling, and the potential significance of ABA-induced actin reorganization is described below.
Signal transduction, transcription, and translation processing
The signal transduction pathway starts with signal perception by different receptors, such as ion channels. In plant cells, the transient influx of Ca2+ ions is one of the earliest responses to salinity stress. The initial increase in cytosolic Ca2+ is followed by the generation of secondary signaling molecules such as inositol phosphates or reactive oxygen species. In turn, secondary signaling molecules can provoke the release of other receptor-mediated Ca2+. The influx of Ca2+ ions initiates the protein phosphorylation cascade, followed by the activation of proteins that are directly involved in cellular protection or the regulation of stress-responsive genes (Xiong et al. 2002).
In our study, we identified four proteins that are directly or implicitly involved in PI signaling: EF1α (spot 15), calreticulin (spot 17), putative calcium-binding protein annexin 6 (spot 22), and 14-3-3-like protein A (spot 33), which were responsive to salinity. EF1α was also altered by ABA pretreatment.
EF1α is a common protein elongation factor, but there is evidence to suggest that it also plays an important role in controlling phosphatidylinositol signaling, which leads to F-actin remodeling (Amiri et al. 2007) and increases the amount of CK surrounding ER and Lys content in maize endosperm (Azama et al. 2003). eEF1α has the ability to bind and bundle F-actin. During PI signaling, PI is converted via PIP and PIP2 forms to IP3, which triggers the release of Ca2+ from cellular stores. Actin MFs could regulate the activity of Ca2+ channels (Wang et al. 2011). It has been reported that PIK-A49, an eEF1A-like protein isolated from carrots, binds and activates carrot phosphatidylinositol-4 kinase (PI4K) in response to an increase in cytosolic calcium. In turn, PI4K catalyzes the biosynthesis of phosphatidylinositol-4 phosphate (PI4P) (Yang and Boss 1994). According to Choi et al. (2008), PIP (PI3P and PI4P) participates in the reorganization of ABA-induced actin filament and stomatal closing in guard cells. They suggested that ABA increases the content of PI3P and PI4P which are involved in the generation of ROS. This triggers an influx of Ca2+, the activation of actin-binding proteins (ABPs), such as profilin, and facilitates the reorganization of actin.
In the presence of calcium, membrane binding proteins, annexins, interact with membrane phospholipids and activate PI signaling. Two proteins were identified in this study: putative calcium-binding protein annexin 6 (spot 22) and 14-3-3A (spot 33) as positive and negative regulators of protein kinase C (PKC), respectively. Annexin A6 acts as a scaffold for recruiting PKCα to the plasma membrane, without its phosphorylation (Lizarbe et al. 2013). 14.3.3A inhibits PKC and activates H+-ATPase (Chen et al. 1994; Chen et al. 2006a, b; Schoonheim et al. 2007). The downregulation of 14.3.3 during salinity stress could result in: (1) reduction of H+-ATPase activity, followed by stomatal closure to prevent water loss and (2) phosphorylation of PKC and activation of Ca2+-ATPase by receptor-mediated hydrolysis of inositol phospholipids.
The Ca2+-storage protein calreticulin is an ER chaperone protein which is also found in Golgi bodies and plasma membrane. Cytosolic Ca2+ levels increase under salt stress when they are released from calcium storage sources. Typically, calreticulin is activated under stress conditions (Tuteja and Sopory 2008). In our study, salinity reduced calreticulin levels, which can be attributed to the release of free Ca2+.
Our study also demonstrated changes in translation and transcription processing in response to salinity. Histone acetyltransferase HAC2 (spot 32) and 60 kDa jasmonate-induced proteins (spot 7) were upregulated during salinity stress. Both HAC2 and 60 kD JIP were ABA-modulated. Acetylation of histones leads decreases transcriptional activation by: (1) DNA relaxation and (2) changes in the interactions between chromatin and chromatin-associated proteins (Fang et al. 2014). Histone acetyltransferases (HATs) are involved in the chromatin-mediated mechanism of stress tolerance in plants exposed to ABA and salt stress (Chen et al. 2006a, b; Sokol et al. 2007; Zhou et al. 2009). 60 kDa jasmonate-induced protein (JIP60) is a ribosome-inactivating protein (RIP). JIP60 exhibited N-glycosidase activity and hydrolyzed the adenine residue from a conserved loop of 25S rRNA (α-sarcin/ricin loop). The α-sarcin/ricin loop binds elongation factors (EFs) to ribosomes. Therefore, any modifications in this fragment of 25S rRNA lead to the inactivation of the large ribosomal subunit and cessation of protein synthesis during the elongation step (Dunaeva et al. 1999; Sawasaki et al. 2008). JIP60 also plays a key role in inhibiting cytoplasmic protein synthesis via reversible ribosome dissociation into small and large subunits (Reinbothe et al. 1994; Rustgi et al. 2014). Some authors have suggested that the upregulation of JIP60 during salinity stress inhibits growth (Görschen et al. 1995) and protects plants against stress. Our results confirm the correlation between salinity and higher levels of 60 kDa jasmonate-induced protein. Surprisingly, exogenously applied ABA inhibited the expression of JIP60 (independent experiment, data not shown). Our findings differ from the observation that ABA, similar to jasmonic acid (JA), can induce JIPs (Lehmann et al. 1995). According to some reports, ABA promotes JA accumulation (Fan et al. 2009). Furthermore, the presence of MYC and NAC transcription factors suggests the existence of cross-talk between JA and ABA signaling (Singh and Laxmi 2015). It should also be noted that ABA and JA antagonistically regulate the expression of salt stress-inducible proteins (Moons et al. 1997). In our previous study (Szypulska and Weidner 2016), the following responses were observed in salt-stressed barley sprouts: (1) increase in the content of free cytosolic polysomes resulting from damage to cell membranes and (2) increase in the concentrations of ribosomal units and free monosomes. The results of the present study suggest that the above could be attributed to the action of JIP60. ABA’s antagonistic effect on JIP60 expression could explain the inhibition of ribosome dissociation and, consequently, the stabilization of polysomes, which was observed in our previous study.
Response to desiccation stress
We identified two main groups of proteins involved in responses to desiccation stress: HSP and LEA proteins. These proteins were upregulated mainly under salinity stress, excluding the HSC70-2 protein (spot 8) which was strongly downregulated.
In general, HSP70 chaperones are involved in the folding of newly synthesized protein chains, prevention of aggregation and refolding of proteins denatured by stress, separation of protein aggregates, translocation of proteins across membranes, and disposal of misfolded proteins (Yong et al. 2003; Cazale et al. 2009; Song et al. 2014). The above explains the high levels of 70 kDa HSPs (spot 6) in stress conditions. The negative effect of salinity on HSC70 (heat-shock protein cognate) expression is unclear, and it could result from low protein yield under stress (Badowiec et al. 2012). HSC70-2 was the only protein whose expression increased under exposure to abscisic acid. The upregulation of HSC70 protein in response to ABA treatment provides additional evidence that translation processes are more effective in response to ABA treatment.
Late embryogenesis-abundant (LEA) proteins and members of this group, such as RAB (responsive to ABA) proteins, also known as dehydrins (DHN), are thought to be involved in protein repair during water stress. Due to their chaperone-like properties, LEA proteins assist proteins that were denatured or misfolded by water stress in recovering their native conformation (Campbell and Close 1997; Brini et al. 2007). In our study, the following LEA proteins were upregulated due to salinity stress: dehydrin (spot 9), dehydrin 6 (spot 18), group 3 LEA proteins (spot 16), late embryogenesis-abundant protein B19.3 (spot 39), protein HVA22 (spot 41), dehydrin DHN1 (spot 34), dehydrin DHN2 (spot 44) and dehydrin DHN3 (spot 42). In this group, HVA22, dehydrin 6, dehydrin DHN2, and dehydrin DHN3 were altered under ABA pretreatment.
Exposure to salinity stress also increased the levels of ABA-inducible protein PHV A1 (spot 35) and salt stress-induced protein (spot 38), both of which were altered by ABA. Hong et al. (1988) demonstrated that the amino acid sequence of PHV A1 shares homology with the LEA7 protein in cotton. Claes et al. (1990) observed that the organ-specific response of sa/T (transcripts of salt stress-induced protein) was correlated with the pattern of Na+ accumulation during salt stress.
Protein turnover
The ubiquitin–proteasome pathway is largely responsible for the degradation of specific cellular proteins during various processes, including exposure to salinity stress (Kornitzer and Ciechanover 2000; Wang et al. 2011; Lyzenga and Stone 2012; Zhang et al. 2014). According to Yan et al. (2000), ubiquitin-specific proteases (USPs)) play a host of important roles in the ubiquitin/26S proteasome pathway by: (1) releasing of free ubiquitins from their translation products, (2) recycling of ubiquitin during degradation of the target compound, which involves the removal of ubiquitin from peptide fragments and splitting of multiubiquitin chains, and/or (3) removing ubiquitins from conjugates before the degradation of the target compound. The latter role indicates that USPs prevent target degradation. These deubiquitinating enzymes (DUBs) are a group of ubiquitin-specific thiol proteases which break the bond between the Gly C-terminus of ubiquitin and covalently attached polypeptides (Yan et al. 2000). In our study, ubiquitin-specific protease 25 (spot 3) was upregulated under salinity stress. The described processes explain the role of protease which was identified in this study in response to salinity stress.
Other defense responses
The following salt-responsive proteins were identified in this study: RNase S-like protein (spot 29), putative endonuclease (spot 30) and trypsin inhibitor CMe (spot 43). All of the above proteins were upregulated in response to salinity and were not modified by ABA pretreatment.
It has been reported that S-like RNase genes are induced in response to various biotic and abiotic factors (Ye and Droste 1996; Galiana et al. 1997; Lee et al. 2009; Zheng et al. 2014). Although the amino acid sequence of S-like RNases is closely related to that of S-RNase, it has no RNase activity (Van Damme et al. 2000; Lee et al. 2009). S-like RNases play an important role in phosphate recycling during senescence and are induced by inorganic phosphate starvation, which is associated with the accumulation of anthocyanins (Ticconi et al. 2001; Lee et al. 2009). Salinity stress induces the accumulation of anthocyanins (Borghesi et al. 2011). Pi starvation could also be associated with salinity stress.
During oxidative stress, the influx of ROS can damage DNA structure (Saha et al. 2015). Efficient DNA repair mechanisms remove oxidized bases. For example, endonuclease III (NTH protein) and endonuclease VIII (NEI protein) from Escherichia coli cells excise many oxidized pyrimidines (Laval 1996; Kumari et al. 2009). The observed upregulation of endonuclease (spot 30) could play a role in DNA repair during salinity stress accompanied by oxidative stress.
Specific protease inhibitors are currently being overexpressed in certain transgenic plants to protect them against invaders (Habib and Fazili 2007) rather than to induce stress tolerance. It has been reported that the sequence of the maize α-amylase/trypsin inhibitor resembles the sequence of osmotin (Claes et al. 1990), a salt-responsive protein. However, protease inhibitors could inhibit the degradation of storage proteins and stilt the growth of salt-stressed seedlings.
Influence of ABA pretreatment on salinity response—summary
In general, ABA pretreatment limited stress-induced changes in the proteome. Interestingly, ABA-inducible protein PHV A1 (spot 35) was one of the five strongly downregulated proteins (to the level comparable with that noted in the control treatment). This finding suggests that the influence of ABA pretreatment on salinity response results from a regulation process, but not downregulation in the strict sense. It implies that ABA plays a major role in plant responses to salinity stress.
The observed changes in GAPDH point to the importance of glycolytic/gluconeogenic pathways in the ABA-modulated salinity response. The regulation of HMT suggests that ABA influences the SMM cycle. The noted changes in the expression of HMT and SHMT indicate that ABA is implicated in the synthesis of osmoprotectants. Other observations demonstrate the important role of ABA in hemicellulose, cellulose, lignin biosynthesis, and ROS scavenging. ABA’s influence on cytoskeletal structure and formation was manifested in the reorganization of actin microfilaments (MFs). We allow for the possibility that GAPDH, in addition to formins the proteins classified to ABPs (actin-binding proteins), can take part in this process. The alterations in EF1α and HAC2 could indicate that ABA participates in the control of PI signaling and chromatin-mediated mechanism of stress tolerance. The number of proteins responsive to desiccation stress was altered by ABA pretreatment (mainly dehydrins), which suggests that ABA contributes to protection against dehydration. Our previous research revealed that ABA treatment increases the concentration of cytomatrix-bound polysomes, a fraction of polysomes that are attached to the membrane and the cytoskeleton, mainly actin microfilaments (Szypulska and Weidner 2011). In that study, we suggested the role of cytomatrix-bound polysomes in LEA proteins synthesis. Moreover, our previous study demonstrated that exogenous abscisic acid stabilizes polysomes, which could be attributed to the high concentration of cytomatrix-bound structures (Weidner et al. 2006). The present study demonstrated the role of ABA in JIP regulation, which expands our knowledge of other factors implicated in mRNA stabilization. In the light of our present and previous findings, we have proposed a hypothetical model of ABA’s role in the induction of salt tolerance in barley sprouts (Fig. 5).
Hypothetical model of ABA’s influence on salt tolerance in higher plants. The role of ABA in salt response may consist in: (1) PI signaling and Ca2+ release due to EF1α regulation and cytoskeletal reorganization, (2) increase in cell wall resistance due to changes in hemicellulose, cellulose, and lignin synthesis, (3) cytoskeleton remodeling due to interactions with ABP and GAPDH, (4) ROS scavenging by antioxidant enzymes (GST, CAT, and POX), (5) chromatin relaxation due to changes in HAC2, (6) translation regulation due to improved mRNA stabilization, (7) dehydration tolerance, mainly due to synthesis of LEA proteins in RER, and (8) osmoprotectant synthesis (DMSP, betaine). The regulated metabolic pathways are marked with bold arrows. ABP actin-binding protein (e.g., formin), cis-regulatory elements: ABRE ABA-responsive element, DRE/CRT dehydration-responsive element/C-repeat, MYBR/MYCR myeloblastosis oncogene/myelocytomatosis oncogene recognition element, NACR non-aggressive challenge responsive element, CK cytoskeleton, CMBP cytoskeleton-membrane-bound polysomes (cytomatrix-bound polysomes), CRT calreticulin, CW cell wall, DAG diacylglycerol, ER endoplasmic reticulum, MF actin microfilament, PM plasma membrane, R potential ABA receptor, and RER rough endoplasmic reticulum. A detailed description can be found in the text
Author contribution statement
ES: idea, realization, laboratory work, and description of results. KJ: laboratory work. SW: financial support and consultancy.
Abbreviations
- 2D PAGE:
-
Two-dimensional electrophoresis in polyacrylamide gel
- ABA:
-
Abscisic acid
- ACTH:
-
Adrenocorticotrophic hormone
- CHAPS:
-
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate
- CHCA:
-
α-Cyano-4-hydroxycinnamic acid
- CK:
-
Cytoskeleton
- DHN:
-
Dehydrins
- DMSP:
-
3-Dimethylsulfoniopropionate
- DTT:
-
Dithiothreitol
- DUBs:
-
Deubiquitinating enzymes
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- GPX:
-
Glutathione peroxidase
- GST:
-
Glutathione S-transferase
- HAC:
-
Histone acetyltransferase
- HATs:
-
Histone acetyltransferases
- HGA:
-
Homogalacturonan
- HMT:
-
Homocysteine methyltransferase
- HSC:
-
Heat shock protein cognate
- HSP:
-
Heat shock protein
- IEF:
-
Isoelectrofocusing
- JIP60:
-
60 kDa jasmonate-induced protein
- LEA proteins:
-
Late embryogenesis-abundant proteins
- MALDI–TOF/MS:
-
Matrix-assisted laser desorption/ionization time-of-flight/mass spectrometry
- MMT:
-
Methionine methyltransferase
- MFs:
-
Microfilaments
- PKC:
-
Protein kinase C
- PMF:
-
Peptide mass fingerprinting
- PMT:
-
Pectin methyltransferase
- ROS:
-
Reactive oxygen species
- SAM:
-
S-Adenosylmethionine
- SMM:
-
S-Methylmethionine
- SHMT:
-
Serine hydroxymethyltransferase
- TFA:
-
Trifluoroacetic acid
- UDPGDH:
-
UDP-d-glucose dehydrogenase
- USPs:
-
Ubiquitin-specific proteases
References
Agarwal S, Sairam RK, Srivastava GC, Meena RC (2005) Changes in antioxidant enzymes activity and oxidative stress by abscisic acid and salicylic acid in wheat genotypes. Biol Plant 49(4):541–550
Aktories K, Lang AE, Schwan C, Mannherz HG (2011) Actin as target for modification by bacterial protein toxins. FEBS J 278:4526–4543
Amiri A, Noei F, Jeganathan S, Kulkarni G, Pinke DE, Lee JM (2007) eEF1A2 activates akt and stimulates akt-dependent actin remodeling, invasion and migration. Oncogene 26:3027–3040
Apraiz I, Mi J, Cristobal S (2006) Identification of proteomic signatures of exposure to marine pollutants in mussels (Mytilus edulis). Mol Cell Proteom 5:1274–1285
Azama K, Abe S, Sugimoto H, Davies E (2003) Lysine-containing proteins in maize endosperm: a major contribution from cytoskeleton-associated carbohydrate-metabolizing enzymes. Planta 217:628–638
Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In: Preiss J (ed) The biochemistry of plants. Academic Press, New York, pp 297–371
Badowiec A, Sylwia Świgońska S, Szypulska E, Weidner S (2012) Influence of exogenous abscisic acid on alterations in protein expression in the proteome of Triticosecale seedlings. Acta Physiol Plant 34:2359–2368
Bakht J, Khan MJ, Shafi M, Khan MA, Sharif M (2012) Effect of salinity and aba application on proline production and yield in wheat genotypes. Pak J Bot 44(3):873–878
Batool N, Shahzad A, Ilyas N, Noor T (2014) Plants and salt stress. Int J Agric Crop Sci 7(9):582–589
Bohnert HJ, Jensen RG (1996) Strategies for engineering water-stress tolerance in plants. Trends Biotechnol 14:89–97
Borghesi E, González-Miret ML, Escudero-Gilete ML, Malorgio MLF, Heredia FJ, Meléndez-Martínez AJ (2011) Effects of salinity stress on carotenoids, anthocyanins, and color of diverse tomato genotypes. J Agric Food Chem 59(21):11676–11682
Brini F, Hanin M, Lumbreras V, Irar S, Pagès M, Masmoudi K (2007) Functional characterization of DHN-5, a dehydrin showing a differential phosphorylation pattern in two Tunisian durum wheat (Triticum durum Desf.) varieties with marked differences in salt and drought tolerance. Plant Sci 172:20–28
Campbell SA, Close TJ (1997) Dehydrins: genes, proteins, and associations with phenotypic traits. New Phytol 137:61–74
Carlier MF, Pantaloni D (2007) Control of actin assembly dynamics in cell motility. J Biol Chem 282:23005–23009
Cazale A-C, Clément M, Chiarenza S, Roncato M-A, Pochon N, Creff A, Marin E, Leonhardt N, Noël LD (2009) Altered expression of cytosolic/nuclear HSC70-1 molecular chaperone affects development and abiotic stress tolerance in Arabidopsis thaliana. J Exp Bot 60(9):2653–2664
Chen Z, Fu H, Liu D, Chang P-FL, Narasimhan M, Ferl R, Hasegawa PM, Bressen RA (1994) A NaCI-regulated plant gene encoding a brain protein homolog that activates ADP ribosyltransferase and inhibits protein kinase C. Plant J 6(5):729–740
Chen F, Li Q, Sun L, He Z (2006a) The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res 13:53–63
Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu J-K, Gong Z (2006b) Mutations in ABO1/ELO2, a subunit of holo-elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol 26(18):6902–6912
Chinnusamy V, Jagendorf A, Zhu J-K (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Choi Y, Lee Y, Jeon BW, Staiger CJ, Lee Y (2008) Phosphatidylinositol 3- and 4-phosphate modulate actin filament reorganization in guard cells of day flower. Plant Cell Environ 31(3):366–377
Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2:19–27
Dunaeva M, Goebel C, Wasternack C, Parthier B, Goerschen E (1999) The jasmonate-induced 60 kDa protein of barley exhibits N-glycosidase activity in vivo. FEBS Lett 452:263–266
Dunand C, Crèvecoeur M, Penel C (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol 174:332–341
Espartero J, Pintor-Toro JA, Pardo JM (1994) Differential accumulation of S-adenosylmethionine synthetase transcripts in response to salt stress. Plant Mol Biol 25(2):217–227
Fahad S, Hussain S, Matloob A, Khan FA, Khaliq A, Saud S, Hassan S, Shan D, Khan F, Ullah N, Faiq M, Khan MR, Tareen AK, Khan A, Ullah A, Ullah N, Huang J (2015) Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul 75(2):391–404
Fan J, Hill L, Crooks C, Doerner P, Lamb C (2009) Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiol 150:1750–1761
Fang H, Liu X, Thorn G, Duan J, Tian L (2014) Expression analysis of histone acetyltransferases in rice under drought stress. Biochem Biophys Res Commun 443:400–405
Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB (2006) Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J 4:145–167
Fatehi F, Hosseinzadeh A, Alizadeh H, Brimavandi T, Struik PC (2012) The proteome response of salt-resistant and salt-sensitive barley. Mol Biol Rep 39:6387–6397
Fujiwara T, Mitsuya S, Miyake H, Hattori T, Takabe T (2010) Characterization of a novel glycinebetaine/proline transporter gene expressed in the mestome sheath and lateral root cap cells in barley. Planta 232:133–143
Fukuda A, Tanaka Y (2006) Effects of ABA, auxin and gibberellin on the expression of genes for vacuolar H+-inorganic pyrophosphatase, H+-ATPase subunit A, and Na+/H+ antiporter in barley. Plant Physiol Biochem 44:351–358
Galiana E, Bonnet P, Conrod S, Keller H, Panabieres F, Ponchet M, Poupet A, Ricci P (1997) RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiol 115:1557–1567
Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002a) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116:238–247
Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002b) Proteomics of Arabidopsis seed germination. A comparative study of wild type and gibberellin deficient seeds. Plant Physiol 129:823–837
Gill SS, Tuteja N (2010) Polyamines and abiotic stress tolerance in plants. Plant Signal Behav 51:26–33
Gimeno-Gilles C, Lelièvre E, Viau L, Malik-Ghulam M, Ricoult C, Niebel A, Leduc N, Limami AM (2009) ABA-mediated inhibition of germination Is related to the inhibition of genes encoding cell-wall biosynthetic and Architecture: modifying enzymes and structural proteins in medicago truncatula embryo axis. Mol Plant 2(1):108–119
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151
Gomez LD, Gilday A, Feil R, Lunn JE, Graham IA (2010) AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64:1–13
Görschen E, Dunaeva M, Hause B, Reeh I, Wasternack C, Parthier B (1995) Expression of the ribosome-inactivating protein JIP60 from barley in transgenic tobacco leads to an abnormal phenotype and alterations on the level of translation. Planta 202:470–478
Goubet F, Mohnen D (1999) Subcellular localization and topology of homogalacturonan methyltransferase in suspension-cultured Nicotiana tabacum cells. Planta 209:112–117
Guan LM, Zhao J, Scandalios JG (2000) Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J 22(2):87–95
Guo CWL, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wanga X (2012) Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the arabidopsis response to stress. Plant Cell 24:2200–2212
Habib H, Fazili KM (2007) Plant protease inhibitors: a defense strategy in plants. Biotechnol Mol Biol 2(3):068–085
Han SJ, Yoo YJ, Kang HS (1995) Characterization of a bifunctional cellulase and its structural gene. J Biol Chem 270(43):26012–26019
Hanson AD, Rivoal J, Paquet L, Gage DA (1994) Biosynthesis of 3-dimethylsulfoniopropionate in Wollastonia biflora (L.) DC. Evidence that S-methylmethionine is an intermediate. Plant Physiol 105:103–110
Hong B, Uknes SJ, T-hD Ho (1988) Cloning and characterization of cDNA encoding a mRNA rapidly-induced by ABA in barley aleurone layers. Plant Mol Biol 11:495–506
Howard J, Hyman AA (2003) Dynamics and mechanics of the microtubule plus end. Nature 422:753–758
Iqbal N, Umar S, Khan NA, Khan MIR (2014) A new perspective of phytohormones in salinity tolerance: regulation of proline metabolism. Environ Exp Bot 100:34–42
Jiang M, Zhang J (2001) Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol 42(11):1265–1273
Jiang M, Zhang J (2003) Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J Exp Bot 53:2401–2410
Jones H, Leigh RA, Tomos AD, Jones RGW (1987) The effect of abscisic acid on cell turgor pressures, solute content and growth of wheat roots. Planta 170(2):257–262
Jung HG, Deetz DA (1993) Cell wall lignification and degradability. In: Jung HG et al (eds) Forage cell wall structure and digestibility. ASA-CSSA-SSSA, Madison, pp 315–346
Kaur N, Reumann S, Hu J (2009) Peroxisome biogenesis and function. Arabidopsis Book 7:e0123
Keskin BC, Sarikaya AT, Yüksel B, Memon AR (2010) Abscisic acid regulated gene expression in bread wheat (Triticum aestivum L.). AJCS 4(8):617–625
Kornitzer D, Ciechanover A (2000) Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol 182:1–11
Kristensen BK, Bloch H, Rasmussen SK (1999) Barley coleoptile peroxidases: purification, molecular cloning and induction by pathogens. Plant Physiol 120:501–512
Krupková E, Immerzeel P, Pauly M, Schmülling T (2007) The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J 50:735–750
Kumari S, Panjabi nee Sabharwal V, Kushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A (2009) Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genom 9(1):109–123
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685
Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62(14):4731–4748
Laval J (1996) Role of DNA repair enzymes in the cellular resistance to oxidative stress. Pathol Biol (Paris) 44(1):14–24
Lee S-H, Lee K-W, Kim K-Y, Choi GJ, Yoon SH, Ji HC, Seo S, Lim YC, Ahsan N (2009) Identification of salt-stress induced differentially expressed genes in barley leaves using the annealing-control-primer-based GeneFishing technique. Afr J Biotechnol 8:1326–1331
Lehmann J, Atzorn R, Brückner C, Reinbothe S, Leopold J, Wasternack C, Parthier B (1995) Accumulation of jasmonate, abscisic acid, specific transcripts and proteins in osmotically stressed barley leaf segments. Planta 197:156–162
Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49:199–222
Lindberg U, Karlsson R, Lassing I, Schutt CE, Hoglund AS (2008) The microfilament system and malignancy. Semin Cancer Biol 18:2–11
Lizarbe MA, Barrasa JI, Olmo N, Gavilanes F, Turnay J (2013) Annexin–phospholipid interactions. Functional implications. Int J Mol Sci 14:2652–2683
Lyzenga WJ, Stone SL (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63:599–616
Mahmood K (2011) Salinity tolerance in barley (Hordeum vulgare L.): effects of varying NaCl, K+/Na+ and NaHCO3 levels on cultivars differing in tolerance. Pak J Bot 43(3):1651–1654
Manjunath S, Sachs MM (1997) Molecular characterization and promoter analysis of the maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family and its expression during anoxia. Plant Mol Biol 33:97–112
Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18:2749–2766
Möller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52:561–591
Moons A, Prinsen E, Bauw G, Montagu MV (1997) Antagonistic effects of abscisic acid and jasmonates on salt stress-lnducible transcripts in rice roots. Plant Cell 9:2243–2259
Neuhoff V, Arold N, Taube D, Ehrhard W (1988) Improved staining in proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9(6):255–262
Ogawa S, Mitsuya S (2012) S-methylmethionine is involved in the salinity tolerance of Arabidopsis thaliana plants at germination and early growth stages. Physiol Plant 144:13–19
Park MY, Kang J-y, Kim SY (2011) Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol Cells 31(5):447–454
Passardi F, Longet D, Penel C, Dunand C (2004a) The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65:1879–1893
Passardi F, Penel C, Dunand C (2004b) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9:534–540
Ranocha P, McNeil SD, Ziemak MJ, Li C, Tarczynski MC, Hanson AD (2001) The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J 25:575–584
Reinbothe S, Reinbothe C, Lehmann J, Becker W, Apel K, Parthier B (1994) JIP60, a methyl jasmonate-induced ribosome-inactivating protein involved in plant stress reactions. Proc Natl Acad Sci 91:7012–7016
Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR (2003) Cell migration: integrating signals from front to back. Science 302:1704–1709
Romero S, Didry D, Larquet E, Boisset N, Pantaloni D, Carlier MF (2007) How ATP hydrolysis controls filament assembly from profilin–actin: implication for formin processivity. J Biol Chem 282:8435–8445
Roxas VP, Smith RK, Allen ER, Allen RD (1997) Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol 15:988–991
Roxas VP, Lodhi SA, Garrett DK, Mahan JR, Allen RD (2000) Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol 41(11):1229–1234
Rustgi S, Pollmann S, Buhr F, Springer A, Reinbothe C, von Wettstein D, Reinbothe S (2014) JIP60-mediated, jasmonate- and senescence-induced molecular switch in translation toward stress and defense protein synthesis. PNAS 111(39):14181–14186
Saha P, Mukherjee A, Biswas AK (2015) Modulation of NaCl induced DNA damage and oxidative stress in mungbean by pretreatment with sublethal dose. Biol Plant 59:139–146
Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidan M, El-Sibai M, Desmarais V, Holman HA, Kitchen S, Backer JM, Alberts A, Condeelies J (2008) WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol 180:1245–1260
Sawasaki T, Nishihara M, Endo Y (2008) RIP and RALyase cleave the sarcin/ricin domain, a critical domain for ribosome function, during senescence of wheat coleoptiles. Biochem Biophys Res Commun 370:561–565
Schoonheim PJ, Sinnige MP, Casaretto JA, Veiga H, Bunney TD, Quatrano RS, de Boer AH (2007) 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J 49:289–301
Shi H, Zhu J-K (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol 50:543–550
Singh D, Laxmi A (2015) Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6:895
Sokol A, Kwiatkowska A, Jerzmanowski A, Prymakowska-Bosak M (2007) Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227:245–254
Song A, Zhu X, Chen F, Gao H, Jiang J, Chen S (2014) A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int J Mol Sci 15:5063–5078
Sripinyowanich S, Klomsakul P, Boonburapong B, Bangyeekhun T, Asami T, Gu H, Buaboocha T, Chadchawan S (2013) Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ Exp Bot 86:94–105
Szyjanowicz PM, McKinnon I, Taylor NG, Gardiner J, Jarvis MC, Turner SR (2004) The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J 37:730–740
Szypulska E, Weidner S (2011) Importance of cytomatrix-bound polysomes to synthesis of lysine-containing proteins in triticale germs under ABA treatment. Acta Physiol Plant 33:1461–1465
Szypulska E, Weidner S (2016) ABA pretreatment can alter the distribution of polysomes in salt-stressed barley sprouts. Acta Mus Siles Sci Nat 65:257–261
Ticconi CA, Delatorre CA, Abel S (2001) Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol 127:963–972
Tuteja N, Sopory SK (2008) Chemical signaling under abiotic stress environment in plants. Plant Signal Behav 3(8):525–536
Van Damme EJM, Hao Q, Barre A, Rouge P, Van Leuven F, Peumans WJ (2000) Major protein of resting rhizomes of Calystegia sepium (hedge bindweed) closely resembles plant RNases but has no enzymatic activity. Plant Physiol 122:433–445
Vannier MP, Thoiron B, Morvan C, Demarty M (1992) Localization of methyltransferase activities throughout the endomembrane system of flax (Linum usitatissimum L.) hypocotyls. Biochem J 286:863–868
Velasco R, Salamini F, Bartels D (1994) Dehydration and ABA increase mRNA levels and enzyme activity of cytosolic GAPDH in the resurrection plant Craterostigma plantagineum. Plant Mol Biol 26:541–546
Verslues PE, Kim YS, Zhu JK (2007) Altered ABA, proline and hydrogen peroxide in an Arabidopsis glutamate:glyoxylate aminotransferase mutant. Plant Mol Biol 64:205–217
Wang S, Kurepa J, Hashimoto T, Smalle JA (2011) Salt stress–induced disassembly of Arabidopsis cortical microtubule arrays involves 26S proteasome-dependent degradation of SPIRAL1 C. Plant Cell 23:3412–3427
Weidner S, Każarnowicz M, Frączek E, Amarowicz R, Karamać M (2006) Exogenous abscisic acid increases stability of polysomes in embryos of triticale caryopses during germination. Acta Physiol Plant 28:627–634
Wibig T (2007) PCA data analysis. Chair of Theoretical Physics, Publishing House of University in Lodz, Lodz
Wilkinson S, Davies WJ (2010) Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ 33:510–525
Wu L, Ge Q, Zhang J, Zhou J, Xu J (2013) Proteomic analysis of Cd-responsive proteins in Solanum torvum. Plant Mol Biol Rep 31:485–491
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14:S165–S183
Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD (2000) The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine1. Plant Physiol 124:1828–1843
Yang W, Boss WF (1994) Regulation of phosphatidylinositol 4-kinase by the protein activator PIK-A49. J Biol Chem 269(5):3852–3857
Yang SS, Zhai QH (2017) Cytosolic GAPDH: a key mediator in redox signal transduction in plants. Biol Plant. doi:10.1007/s10535-017-0706-y
Ye Z-H, Droste DL (1996) Isolation and characterization of cDNAs encoding xylogenesis-associated and wounding-induced ribonucleases in Zinnia elegans. Plant Mol Biol 30:697–709
Yong JC, Barral JM, Ulrich Hartl F (2003) More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci 28(10):541–547
Yu L (2008) Cell behavior and the role of profilin. Cell Biol 978-91-7155-677-6:1-53
Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song CP (2001) Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol 126(4):1438–1448
Zhang A, Jiang M, Zhang J, Ding H, Xu S, Hu X, Tan M (2007) Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol 175(1):36–50
Zhang Y, Cheng Y, Guo J, Yang E, Liu C, Zheng X, Deng K, Zhou J (2014) Comparative transcriptome analysis to reveal genes involved in wheat hybrid necrosis. Int J Mol Sci 15:23332–23344
Zhao Y, Pan Z, Zhang Y, Qu X, Zhang Y, Yang Y, Jiang X, Huang S, Yuan M, Schumaker KS, Guo Y (2013) The actin-related Protein2/3 complex regulates mitochondrial-associated calcium signaling during salt stress in Arabidopsis. Plant Cell 25:4544–4559
Zheng J, Wang Y, He Y, Zhou J, Li Y, Liu Q, Xie X (2014) Overexpression of an S-like ribonuclease gene, OsRNS4, confers enhanced tolerance to high salinity and hyposensitivity to phytochrome-mediated light signals in rice. Plant Sci 214:99–105
Zhou X, Hua D, Chen Z, Zhou Z, Gong Z (2009) Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J 60:79–90
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Hajduch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Szypulska, E., Jankowski, K. & Weidner, S. ABA pretreatment can limit salinity-induced proteome changes in growing barley sprouts. Acta Physiol Plant 39, 190 (2017). https://doi.org/10.1007/s11738-017-2490-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2490-x