Abstract
Background
Nsa cytoplasmic male sterility (CMS) is a novel alloplasmic male sterility system derived from somatic hybridization between Brassica napus and Sinapis arvensis. Identification of the CMS-associated gene is a prerequisite for a better understanding of the origin and molecular mechanism of this CMS. With the development of genome sequencing technology, organelle genomes of Nsa CMS line and its maintainer line were sequenced by pyro-sequencing technology, and comparative analysis of the organelle genomes was carried out to characterize the organelle genome composition of Nsa CMS as well as to identify the candidate Nsa CMS-associated genes.
Results
Nsa CMS mitochondrial genome showed a higher collinearity with that of S. arvensis than B. napus, indicating that Nsa CMS mitochondrial genome was mainly derived from S. arvensis. However, mitochondrial genome recombination of parental lines was clearly detected. In contrast, the chloroplast genome of Nsa CMS was highly collinear with its B. napus parent, without any evidence of recombination of the two parental chloroplast genomes or integration from S. arvensis. There were 16 open reading frames (ORFs) specifically existed in Nsa CMS mitochondrial genome, which could not be identified in the maintainer line. Among them, three ORFs (orf224, orf309, orf346) possessing chimeric and transmembrane structure are most likely to be the candidate CMS genes. Sequences of all three candidate CMS genes in Nsa CMS line were found to be 100% identical with those from S. arvensis mitochondrial genome. Phylogenetic and homologous analysis showed that all the mitochondrial genes were highly conserved during evolution.
Conclusions
Nsa CMS contains a recombined mitochondrial genome of its two parental species with the majority form S. arvensis. Three candidate Nsa CMS genes were identified and proven to be derived from S. arvensis other than recombination of its two parental species. Further functional study of the candidate genes will help to identify the gene responsible for the CMS and the underlying molecular mechanism.
Similar content being viewed by others
Background
Cytoplasmic male sterility (CMS) is a widely spread phenomenon in which the plant is unable to produce functional pollen [1, 2]. This phenomenon exists extensively in plant kingdom as a result of natural variations that follow the evolutionary path of mitochondrial genome rearrangement [3, 4]. CMS caused by natural variation of mitochondrial genome is usually called homoplasmic, which has been found in many crop species, such as WA CMS in rice [5], CMS-T in maize [6], Don CMS in radish [7], Pol CMS [8] and Nap CMS in rapeseed [9]. Some of the homoplasmic CMS has been successfully used as pollination control system for heterosis application, such as Pol CMS in rapeseed. However, drawbacks such as thermo-sensitive nature or lack of restoration in homoplasmic CMS systems have made the exploitation of alloplasmic CMS system desirable.
Interspecific or intergeneric hybridization has been proven efficient for the establishment of alloplasmic CMS in crops via either sexual or somatic hybridization, such as Ogu CMS derived from intergeneric hybridization of Raphanus sativus and Brassica napus [10], and Tour CMS from sexual hybridization of Brassica juncea and Brassica tournefortii [11]. Somatic hybridization is more advantageous compared to sexual hybridization for crossing genetically distant species by overcoming compatibility barrier as well as combining both organelle and nuclear genomes. Studies have revealed that alloplasmic CMS derived from cybrids retains a lot of donor mitochondrial genome sequences, as observed in Solanaceae [12] and Brassicaceae [13,14,15]. However, the composition of organelle genomes at whole genome level of a cybrid-derived alloplasmic CMS and the origin of its sterility gene have not been completely elucidated.
Previous studies have shown that plant CMS genes usually encode a protein with transmembrane domains and have a chimeric structure, containing coding sequences of known genes, e.g. those encoding ATPase, cytochrome-c oxidase, or ribosomal proteins etc. [16,17,18]. The most common method to identify the CMS-associated gene is to compare the differences of mitochondrial genes between the male sterile line and its maintainer lines at genomic, transcriptional and proteomic levels. For example, orf288 as a specific novel chimeric ORF was identified to be Hau CMS candidate gene in B. juncea by comparison of genomic sequence and gene expression between the CMS line and its maintainer line using genome walking and Northern-blot analysis [19]. OrfH79 in rice HL CMS [20] and orf263 in rapeseed Tour CMS [11] were identified by using a similar approach. Other CMS genes, such as orf138 of Ogu CMS in B. napus [21], orf522 in sunflower CMS [22], urf13 of CMS-T in maize [23] were detected by comparing the translated proteins in the CMS and fertile lines.
The advantage of next-generation sequencing (NGS) technology has allowed for convenient identification of CMS gene and a comprehensive investigation of all genes from both chloroplast and mitochondrial genomes. The size of plant chloroplast and mitochondrial genome is usually 120–160 kb and 200–2400 kb, respectively [24,25,26], both are much smaller than nuclear genomes. To date, there has been 3295 and 290 plant species with whole chloroplast or mitochondrial genomes sequenced, respectively (https://www.ncbi.nlm.nih.gov/genome/browse#!/organelles/). Compared to chloroplast genome, the mitochondrial genome is more unstable, with more long, short and tandem repeats, and thus is amenable to rearrangements. By using the whole genome sequencing approach, CMS-associated ORFs, including orf463 in radish [7] and orf507 in pepper [27] were identified.
Oilseed rape (Brassica napus L.) is the third largest oilseed crop worldwide, contributing about 27% of total edible plant oil production [28]. Significant heterosis has dramatically increased the yield of oilseed rape and will continue to contribute to the oilseed industry. CMS as the main pollination control system has been used in oilseed rape hybrid production for more than 20 years, including homoplasmic Pol CMS and alloplasmic Ogu CMS. With the development of embryo rescue and somatic hybridization technology, many alloplasmic CMS have been established [29,30,31,32]. Up to now, there are at least 30 alloplasmic male sterile systems in Brassicaceae crops, which greatly enrich the sterile cytoplasm type [33]. In B. napus, however, only Pol CMS and Ogu CMS are commonly used with some problems such as unstable sterility in Pol CMS and genetic drag of Radish segment in Ogu CMS restorers. Nsa CMS was identified to be a novel B. napus alloplasmic male sterility system derived from somatic hybridization of B. napus cv. Zhongshuang 4 and Sinapis arvensis var. Yeyou 18 [34]. The Nsa CMS line is essentially different from other rapeseed CMS systems such as Ogu, Nap, Pol, Tour and Hau, based on their origins, genetic, morphological, cytological and molecular characterization [35,36,37]. Compared with the most commonly used Pol CMS, Nsa CMS is less susceptible to temperature with stable sterility [29]. Progress has been made on hybrid variety development with Nsa CMS and several new hybrids are on the way for registration. In order to characterize the organelle genome composition and identify candidate CMS-associated genes of Nsa CMS, we sequenced the complete organelle genomes of Nsa CMS line and its iso-nuclear maintainer line of B. napus cv. Zhongshuang 4 which is the original parental line used in somatic hybridization by employing Roche/454 pyro-sequencing technology. Comparative analysis of the organelle genome sequences of the two lines together with Sinapis arvensis var. Yeyou 18, which is the other parental species for Nsa CMS, resulted in the identification of candidate Nsa CMS-associated gene and the composition of organelle genomes in Nsa CMS. Our results will lay a foundation for elucidating the mechanisms of plant CMS and give insights into the formation of alloplasmic male sterility system.
Results
Assembling of chloroplast genomes
The Roche 454 sequencing platform generated 46.3 M bp and 33.4 M bp of sequence reads for Nsa CMS and Zhongshuang 4, respectively. In addition, 546,642 and 618,772 clean reads were generated for Zhongshuang 4 and Nsa CMS by Miseq sequencing. The reads were firstly assembled into 5 contigs in each line with the coverage of 38.8× and 52.3×, respectively. PCR amplification of contig ends showed that there was no missing base between the contigs, and the break between contigs was caused by inverted repeat (IR) sequence. The chloroplast genomes of Nsa CMS and maintainer lines were individually assembled into a single, circular mapping molecule with a size of 153,449 bp (Genbank no. MN428073), and 153,458 bp (Genbank no. MN428074), and G + C content of 36.37 and 36.35%, respectively.
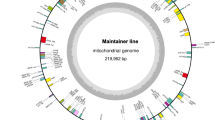
The chloroplast genomes of both lines have similar structure, containing a pair of inverted repeat (IR) regions, which were divided by short single copy (SSC) and long single copy (LSC) regions (Fig. 1). The chloroplast genome possesses a total of 129 genes, including 4 rRNA genes, 30 tRNA genes, and 79 protein-coding genes (Table 1). Within the IR regions, there are 16 duplicate genes, and rps19 gene spans the IR and SSC regions.
Assembling of mitochondrial genomes
The reads data generated from mitochondrial DNA of Nsa CMS and Zhongshuang 4 were assembled into 16 and 9 contigs with the coverage depth of 191× and 195×, respectively. The average contig length of Nsa CMS and Zhongshuang 4 was 15.56 kb and 24.53 kb, respectively. The gap length of Nsa CMS genome between contigs was 0–500 bp, and that of Zhongshuang 4 genome was all less than 50 bp.
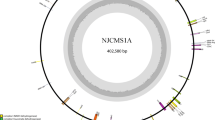
After filling in the gaps, the mitochondrial genomes of Nsa CMS and its maintainer were each assembled into a single, circular mapping molecule with a size of 269,977 bp (Genbank no. MN443182) and 221,862 bp (Genbank no. MN428072), and G + C content of 45.08 and 45.18%, respectively (Fig. 2). The two mitochondrial genomes shared similarities on gene composition, both consisting of 34 protein coding genes, 18 tRNA and 3 rRNA genes (Table 2).
Sequence alignment showed that the mitochondrial genome of Zhongshuang 4 was 9 bp larger than previously reported B. napus cv. “Westar” (Genbank accession number: AP006444.1), and only 31 single nucleotide polymorphisms (SNPs) were detected between the two mitochondrial genomes. One 2427 bp forward repeat sequence was found in mitochondrial genome of Zhongshuang 4, which means a tripartite mitochondrial genomic structure existed in this mitotype [38]. Based on the repeat sequence in the mitotypes, the mitochondrial is inferred as containing one master circle accompanied by two smaller circles of 124,910 bp and 69,952 bp in size, which was similar with Westar [39].
Comparison of protein-coding gene in the mitochondrial genome
Functional genes are conserved in the mitochondrial genome and their evolution is very slow among different plant species [40, 41]. The differences of protein sequences are important candidates for functional analysis of CMS-associated genes as well as revealing the molecular mechanisms of CMS. Based on our sequence data, most protein-coding genes in the mitochondrial genome of B. napus and S. arvensis are identical, especially in size, except rps3 and cox2–2 (S, B, Nsa is prefixed to the names of genes/ORFs in S. arvensis, B. napus and Nsa CMS, respectively). For instance, in the 2nd exon of Brps3, there is a 33 bp insertion in Srps3 and Nsarps3, but not in Nrps3. Another case is cox2–2, which is absent in S. arvensis but present in both B. napus and Nsa CMS.
In addition, there were 26 single nucleotide polymorphism (SNPs) scattered among 12 protein-coding genes. Among these SNPs, 10 were synonymous (nad3, rps12, rps7, orfX, cox1, atp1) and 16 were non-synonymous (Table 3). Most of these SNPs were transitions.
Composition of Nsa CMS organelle genome
To elucidate the formation and organelle composition of Nsa CMS system that established by somatic hybridization, we performed a comparative analysis of the whole organelle genome of Nsa CMS and its parental lines, Zhongshuang 4 (maintainer line) and S. arvensis. The organelle genome of S. arvensis was previously sequenced (Genbank no. KM851044). Although Nsa CMS mitochondrial genome showed some collinearity with the maintainer line, the collinear segments were short and scattered in the genome (Fig. 3). A much higher collinearity of Nsa CMS mitochondrial genome with that of S. arvensis was inferred. Sequence alignment using BLASTN showed that the sequence coverage of Zhongshuang 4 mitochondrial genome to Nsa CMS mitochondrial genome was 86%, with an identity of 99.85%. In contrast, the sequence coverage of S. arvensis mitochondrial genome to that of Nsa CMS was 93%, with an identity of 99.93%.
Comparative diagrams of Nsa CMS, Zhongshuang 4 and Sinapis arvensis mitochondrial genomes. Different blocks are assigned with different color in each mitochondrial genome, and the corresponding line that connects two blocks indicates high homology of these two blocks. Direct or reverse transcript orientation is indicated above and below the central line, respectively
We also compared coding sequences of the mitochondrial genome of the three materials (Table 4). Except for 16 genes which were completely consistent among these three materials, there were 18 coding genes that were different. Among the different genes, 13 were from S. arvensis and 5 genes were from B. napus. In addition, the sequence of rps3 and cox2–2 are different in the mitochondrial genomes of B. napus and S. arvensis, and the deletion in rps3 and cox2–2 were present in Nsa CMS, which means that Nsa CMS has rps3 gene from S. arvensis cv. “Yeyou 18” and cox2–2 gene from Zhongshuang 4. To verify these genomic differences, we designed two primer pairs specific for rps3 and cox2–2, respectively. PCR amplification of rps3 gene showed that an identical fragment was generated from Nsa CMS and S. arvensis, but a shorter fragment from Zhongshuang 4. Meanwhile, an identical fragment of cox2–2 gene was amplified from Nsa CMS and Zhongshuang 4, but no product was obtained from S. arvensis (Fig. 4). Sequencing of the amplified fragments confirmed the deletion in rps3 gene from Nsa CMS and S. arvensis, and the existence of cox2–2 in Nsa CMS and Zhongshuang 4.
Four sequences with 9432 bp, 7383 bp, 2427 bp and 1592 bp length were present in Nsa CMS mitochondrial genome as repeat sequences, with the first three were reverse repeats and the last one is forward repeats. However, repeats with 7383 bp and 2427 bp length appeared as forward repeats in both S. arvensis and Zhongshuang 4 mitochondrial genomes. The 9432 bp and 7383 bp length repeats contained two short repeat sequences, indicating that the inverted repeats can be easily converted into another direction. More repeat sequences may be the reason for the greater number of gaps in the mitochondrial genome of Nsa CMS than those of B. napus and S. arvensis.
Comparative analysis of chloroplast genome showed that Nsa CMS had a higher collinearity with Zhongshuang 4 than S. arvensis. Sequence alignment showed that the identity between chloroplast genomes of Nsa CMS and Zhongshuang 4 was 99.96%, and those of Nsa CMS and S. arvensis was 97.6%, possessing very high collinearity (Additional file 1). Apart from 60 SNPs and 50 gaps (0–5 bp delations or insertions), the chloroplast genome of Nsa CMS was very similar to that of the maintainer line without any structural difference. In contrast, 521 SNPs and 632 gaps between the chloroplast genomes of Nsa CMS and S. arvensis were identified. These results indicate that the chloroplast genome of Nsa CMS was derived directly from its B. napus parent Zhongshuang 4.
Identification of CMS-associated ORFs
Forty-nine ORFs encoding proteins with over 100 amino acids have been found in Nsa CMS mitochondrial genome. Among them, 16 ORFs were specifically detected in Nsa CMS but not in the maintainer line. Transmembrane prediction revealed that 11 of the Nsa CMS specific ORFs possessed transmembrane domains (Table 5, Fig. 5). Three of the orfs, orf224, orf309 and orf346 contained a chimeric structure. The orf224 is a 675 bp gene containing a 175 bp homologous fragment with 98% identity to atp8. Both orf309 and orf346 are chimeric with cox1, in which a 133 bp sequence was found with 98% identity.
Almost all identified plant CMS-associated ORFs were chimeric genes, mostly located upstream or downstream functional genes, encoding components of the electron transport respiratory chain, and possessed transmembrane domains [18]. According to the mitochondrial genome sequencing data, orf224, orf346 and orf309 are all chimeric genes, of which orf224 and orf346 located upstream known genes in Nsa CMS (Fig. 6). Orf346 is located upstream nad3 and rps12, with only 103 bp distance from nad3. ccmB is located 217 bp downstream orf224, but there is no gene located near orf309. All the three orfs existed in S. arvensis but not in the maintainer.
Orf224 in Nsa CMS (Nsaorf224) has 93% sequence identity to that of Pol CMS associated gene orf224 (Polorf224) and there are 27 different amino acids between them. A corresponding orf222 in the maintainer line which is also chimeric with atp8, was found to have 88% similarity with Nsaorf224, and 84% similarity with Polorf224. Orf286 in the maintainer line is homologous to orf309, with 96% sequence similarity. Orf322 in the maintainer line is homologous to orf346 with 91% sequence similarity.
Blast search for homologous of the three orfs from Genbank resulted in 6, 9 and 8 homologous for Nsaorf224, Nsaorf309 and Nsaorf346, respectively. Phylogenetic analysis showed that the relatedness between these three orfs and the homologous from B. napus was quite distant. Nsaorf224 was not grouped with any of the homologous in Brassica species. Orf309 and orf346 showed a closer genetic relationship with homologous in R. sativus than those in Brassica species, and homologous sequence analysis shows that the chimeric structure of cox1 in orf309 and orf346 only existed in R. sativus among Brassicacea species (Fig. 7).
Discussion
Origin of Nsa CMS organelle genome
There has been a large number of study on cytoplasmic source of alloplasmic CMS in plants [12, 42,43,44]. Plenty of work has been done on the analysis of heterologous cytoplasm, but only a few studies reported on the origin of the cytoplasmic genome of heterologous CMS line. For example, the mitochondrial of the first alloplasmic male sterility system in tobacco were confirmed to be a fusion type, whereas the chloroplasts came from a single parent [45, 46]. Alloplasmic male sterility system derived from somatic hybridization between B. napus and A. thaliana contained a mixed mitochondrial composition from both parents [42]. The SW18 mitochondrial genome has the whole circular genome of B. napus cv. “Westar” with about 60% redundancy, and four partial fragments of Kosena radish [30]. Mitochondrial genome sequencing showed that the exogenous sequence was mostly derived from the donor parent (radish) of somatic hybrids in Ogu CMS [44]. Alloplasmic CMS line of durum wheat contained several heterologous regions [47]. From the perspective of current research, plants regenerated from fused cells and their progeny usually inherited the chloroplasts from only one of the parents, while the mitochondrial genome is recombined from both parents [12, 30, 44, 48].
As a novel alloplasmic CMS system developed by somatic hybridization between B. napus and S. arvensis [34], Nsa cytoplasm was proven to be different from other CMS systems in B. napus and contains mitochondrial genomic fragments from both parental species by RFLP markers [36]. In this study, by comparative analysis of the whole organelle genome, we found that Nsa CMS chloroplast genome was completely derived from the B. napus parent, and its mitochondrial genome was derived from the recombination of B. napus and S. arvensis. However, the higher collinearity of Nsa CMS mitochondrial genome with S. arvensis revealed that the majority of Nsa CMS mitochondrial genome was derived from S. arvensis. This is consistent with previous conclusion that cybrid progeny inherit chloroplasts from only one of the parents, while the mitochondrial genome is recombined from both parents [12, 30]. A large number of repeat sequences in mitochondria are the source of mitochondrial recombination, whereas there are much fewer repeat sequences in the chloroplast genome. This might be the reason that the chloroplast can inherit steadily and the mitochondrial genomes undergo recombination in the process of somatic cell fusion.
NGS is an efficient approach to acquire CMS-associated gene
Comparative analysis of the CMS line with its maintainer line on mitochondrial gene structure, expression and translation products has become the conventional approach to identify CMS gene. For example, using genomic walking and Northern-blot analysis, the Hau CMS orf288 in B. juncea [19], orfH79 in rice HL CMS [20] and orf263 in rapeseed Tour CMS [11] were identified. Analysis of protein difference has also led to the identification of orf138 from B. napus Ogu CMS [21], orf522 from sunflower CMS [22], urf13 of CMS-T from maize [23] and orf147 from Cajanus cajan A4 CMS [49].
In recent years, with the development of genome sequencing technology, many CMS genes were identified by directly sequencing and comparison the whole mitochondrial genome of CMS and its maintainer lines, such as orf182 from rice D1 CMS [50], orf507 from peppers [27], orf113 from rice RT98A CMS [51], and orf463 in radish [7]. This approach is much easier and straighter forward than the conventional ones due to the small size of plant mitochondrial genome, which varied from 200 to 2400 kb [44, 52, 53]. In this study, we identified three ORFs specific to the CMS line with transmembrane domain and chimeric structure. All of them contained the typical structure of plant CMS genes and are very likely to be the candidate CMS genes for Nsa CMS.
The origin of Nsa CMS gene
As there is a large number of repetitive sequences in the mitochondrial genome, rearrangement or recombination events of the genome usually occur during species evolution [47]. The instability of mitochondrial genome has been reported the reason for generation of new ORFs, especially those related to CMS [3, 26]. Although, somatic hybridization could accelerate the process of recombination in mitochondrial genome and Nsa CMS mitochondrial genome derived from recombination events between S. arvensis and B. napus, three candidate sterile genes identified were not formed by recombination of the two parental species in this study. All the three candidate genes were directly derived from S. arvensis mitochondrial genome without any sequence variation.
Previous studies showed that genes related to crop CMS were mainly derived from alien or ancestral species, such as orf138 in B. napus from R. sativus, orfH79 and WA352 in O. sativa from wild rice [54, 55],, orf263 in B. juncea from B. tournefortii [11]. Even in CMS systems derived from cybrids, the source of CMS genes was proven to be directly from the donor parental species, such as Brassica CMS genes orf138 and orf125, which originated from radish, orf263 in B. juncea from B. tournefortii [11, 30, 44].
To date, almost all identified CMS-associated ORFs possessed chimeric genes and transmembrane domains [18]. Three Nsa CMS-associated ORFs also possess chimeric structure. However, there are few homologous genes with Nsa CMS-associated genes. It signifies that the mitochondrial genes have considerable stability during evolution and alloplasmic male sterile is caused by incompatibility between cytoplasm and nucleus. The candidate CMS genes identified in our study should also be a result of mitochondrial genome rearrangement, which occurred in S. arvensis or its ancestral species, other than recombination of parental genomes in cybrids.
Structure of CMS gene and its functional prediction
We conducted extensive sequence comparison between B. napus and Nsa CMS mitochondrial genomes to search for functional alterations of genes that were responsible for the CMS phenotype. It is noted that several partial subunits of the respiratory chain complex and cytochrome c assembly protein, including ATP1, NAD4, CCMFN1, CCMFN2 encoded by mitochondrial genome were different between Nsa CMS and its maintainer line. Any of these altered proteins may interfere with the electron transfer chain (mtETC), weakening energy supplies and stalling pollen development [56]. We also observed amino acid variations among RPL5, ORFX (TATC), RPL2, MATR, RPS3 coded by Nsa CMS mitochondrial genome. Almost all the mitochondrial functional genes play an absolutely necessary role in maintaining the completeness of mitochondria and metabolism, and there have not been reports of CMS caused by mutation of functional mitochondrial genes so far. Thus, we think that these differences on functional genes may not directly associate with cytoplasmic male sterility. Whether these variant genes are related to the sterility regulation in Nsa CMS requires further study.
To date, almost all identified CMS genes are novel chimeric open reading frames (ORFs). Studies have shown that at least 10 mitochondrial genes, involving electron transfer chain (mtETC) pathways, such as cox1, atp8, and atp6 were frequently involved in the formation of CMS genes [18]. In rapeseed, orf222 for CMS-Nap and orf224 for CMS-Pol encode proteins with an N terminus similar to ATP8 and the remaining sequence of unknown origin [8, 9]. In Pepper CMS-Peterson and Sugar beet CMS-Owen, the chimeric orf456 and preSatp6 encode a protein with a segment of ATP6 at the N terminus [57, 58]. Rice orf79 for CMS-BT and orfH79 for CMS-HL [59, 60], and radish orf463 for CMS-Don [7] encode small proteins with an N terminus similar to COX1 and the remaining sequence of unknown origin. These CMS genes consisting of portions of some essential mitochondrial genes may competitively interact with the mtETC complexes, and result in decreased ATP production [18].
Also, all the proteins encoded by plant CMS genes possess transmembrane domains, such as URF13 for maize CMS-T [61], ORF138 for B. napus CMS-Ogu [21], ORF79 for rice CMS-BT [60], ORFH79 for rice CMS-HL [59], and ORF224 for rapeseed CMS-Pol [8]. The CMS proteins may integrate into the inner mitochondrial membrane, destroying the proton gradient and affecting ATP synthesis. For example, ORFH79 in HL CMS, interacts with subunit P61 of mtETC complex III, impairing the activity of complex III and resulting in decreased ATP production [59]. This explains the necessity of the sterility proteins being membrane proteins, otherwise they are unable to interfere with energy synthesis.
In the present study, three ORFs with transmembrane and chimeric structures were identified in the mitochondrial genome of Nsa CMS. Of which orf224 is chimeric with atp8, and both orf309 and orf346 are chimeric with cox1. The chimeric segments may interact with corresponding functional genes involved in mtETC leading to fertility abortion of pollen. Therefore, these three ORFs are excellent candidate sterility genes of Nsa CMS. Further functional studies of these candidate genes are required to identify Nsa CMS associated gene and understand the sterility regulation mechanism in Nsa CMS.
Conclusions
The complete organelle genome sequences of Nsa CMS and its maintainer lines in B. napus were obtained and characterized. Comparative analysis of whole organelle genomes of Nsa CMS line and its parents showed that Nsa CMS mitochondrial genome was derived from the asymmetrical fusion of its parent lines, but Nsa CMS chloroplast genome was derived from the B. napus parent. Three ORFs, orf224, orf309 and orf346, coding for hypothetical proteins with a chimeric structure and transmembrane domain were identified as the candidate genes for Nsa CMS. Their existence in both Nsa CMS and S. arvensis revealed that the candidate CMS genes were not caused by rearrangement of mitochondrial genome during somatic hybridization. All of the candidate sterile ORFs were derived from S. arvensis and formed either in S. arvensis or its progenitor.
Materials and methods
Plant materials
Nsa CMS line was produced by repeatedly backcrossing cybrids from somatic hybridization of B. napus cv. “Zhongshuang 4” and S. arvensis cv. “Yeyou 18” with Zhongshuang 4. Seeds of Nsa CMS, Zhongshuang 4 and S. arvensis were harvested from the experimental field of Oil Crops Research Institute, Chinese Academy of Agricultural Sciences in Wuchang, Hubei.
Isolation of organelle DNA
Total DNA was isolated from the organelles by employing discontinuous sucrose gradient as described by Chen et al. (2011) [38]. Seven days old etiolated seedlings grown at 25 °C were used to extract Organelle DNA. CTAB method was employed for total DNA extraction.
Cytoplasmic genome composition identification
The primers were designed according to the sequence differences of rps3 and cox2–2 genes in Nsa CMS and maintainer lines. The primer sequences were listed in Additional file 2. PCR program was as following: 94 °C 30 s, 56 °C 30 s, 72 °C 1 min, 32 circles.
Genome sequencing and sequence analysis
DNA samples with good quality were used to construct sequencing libraries. As Roche 454 Sequencing Platform has a high base error rate, organelle genomes were also sequenced using the Roche 454 FLX + and Illumina MiSeq platforms (Personal Biotechnology, Shanghai, China). Genome sequences were assembled by Newbler Assembler Software Version 2.8 (454 Life Sciences, Branford, USA). Contig gaps were filled by sequences obtained from Sanger sequencing of PCR products amplified with primers designed based on contig end sequences.
BLASTX, BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and tRNA-SE [62] were used to identify mitochondrial and chloroplast rRNA, tRNA and genes. ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to screen hypothetical proteins longer than 100 amino acids by default parameter. Blast2seq (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to perform sequence alignment to find the syntenic region of mitochondrial genomes. The presence of transmembrane domain in each hypothetical protein was performed with online TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Phylogenetic analysis of candidate CMS-associated genes in Brassicaceae was performed by MEGA 7.0 [63]. DOGMA was used to visualize data and information of the mitochondrial genome of Nsa CMS line and its maintainer line [64]. Progressive Mauve was used for multiple alignments among the mitochondrial genomes in Brassica [65]. The Genbank accession numbers of protein sequences involved in this study are listed in Additional file 3.
Availability of data and materials
The datasets generated during the current study are available in the Genbank with accession numbers MN443182 and MN428072 (mitochondrial) and MN428073 and MN428074 (chloroplast) genome sequences, respectively. The Genbank accession number of proteins and genomes used in the study are listed in the Additional file 2 and Additional file 3.
Abbreviations
- CMS:
-
Cytoplasmic male sterility
- IR:
-
Inverted repeat
- LSC:
-
Long single copy
- ML:
-
Maximum likelihood
- NGS:
-
Next-generation sequencing
- ORFs:
-
Open reading frames
- SNPs:
-
Single nucleotide polymorphism site
- SSC:
-
Short single copy
References
Laser KD, Lersten NR. Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot Rev. 1972;38(3):425–54.
An H, Yang Z, Yi B, Wen J, Shen J, Tu J, Ma C, Fu T. Comparative transcript profiling of the fertile and sterile flower buds of pol CMS in B. napus. BMC Genomics. 2014;15(1):258.
Feng X, Kaur A, Mackenzie SA, Dweikat IM. Substoichiometric shifting in the fertility reversion of cytoplasmic male sterile pearl millet. Theor Appl Genet. 2009;118(7):1361–70.
Kubo T, Kitazaki K, Matsunaga M, Kagami H, Mikami T. Male sterility-inducing mitochondrial genomes: how do they differ? Crit Rev Plant Sci. 2011;30(4):378–400.
Virmani S, Wan B. Development of CMS lines in hybrid rice breeding. Hybrid rice. Manila, Philippines: International Rice Research Institute; 1988. p. 103–14.
Levings C 3rd. Thoughts on cytoplasmic male sterility in cms-T maize. Plant Cell. 1993;5:1285.
Park JY, Lee YP, Lee J, Choi BS, Kim S, Yang TJ. Complete mitochondrial genome sequence and identification of a candidate gene responsible for cytoplasmic male sterility in radish (Raphanus sativus L.) containing DCGMS cytoplasm. Theoretical and applied genetics. Genet. 2013;126(7):1763–74.
Handa H, Gualberto JM, Grienenberger JM. Characterization of the mitochondrial orfB gene and its derivative, orf224, a chimeric open reading frame specific to one mitochondrial genome of the “Polima” male-sterile cytoplasm in rapeseed (Brassica napus L.). Curr Genet. 1995;28(6):546–52.
L’Homme Y, Stahl RJ, Li XQ, Hameed A, Brown GG. Brassica nap cytoplasmic male sterility is associated with expression of a mtDNA region containing a chimeric gene similar to the pol CMS-associated orf224 gene. Curr Genet 1997; 31(4): 325–335.
Tanaka Y, Tsuda M, Yasumoto K, Yamagishi H, Terachi T. A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.). BMC Genomics. 2012;13:352.
Landgren M, Zetterstrand M, Sundberg E, Glimelius K. Alloplasmic male-sterile Brassica lines containing B. tournefortii mitochondria express an ORF 3′ of the atp6 gene and a 32 kDa protein. Plant Mol Biol. 1996;32(5):879–90.
Sanchez-Puerta MV, Zubko MK, Palmer JD. Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 2015;206(1):381–96.
Leino M, Landgren M, Glimelius K. Alloplasmic effects on mitochondrial transcriptional activity and RNA turnover result in accumulated transcripts of Arabidopsis orfs in cytoplasmic male-sterile Brassica napus. Plant J. 2005;42(4):469–80.
Du K, Liu Q, Wu X, Jiang J, Wu J, Fang Y, Li A, Wang Y. Morphological structure and transcriptome comparison of the cytoplasmic male sterility line in Brassica napus (SaNa-1A) derived from somatic hybridization and its maintainer line SaNa-1B. Front Plant Sci. 2016;7:1–13.
Kang L, Li P, Wang A, Ge X, Li Z. A novel cytoplasmic male sterility in Brassica napus (inap CMS) with Carpelloid stamens via protoplast fusion with Chinese Woad. Front Plant Sci. 2017;8:529.
Cui X, Wise RP, Schnable PS. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science. 1996;272(5266):1334–6.
Menassa RHY, L’Homme Y, Brown GG. Post-transcriptional and developmental regulation of a CMS-associated mitochondrial gene region by a nuclear restorer gene. Plant J 2010;17(5):491–499.
Chen L, Liu YG. Male sterility and fertility restoration in crops. Annu Rev Plant Biol. 2014;65(1):579–606.
Jing B, Heng S, Tong D, Wan Z, Fu T, Tu J, Ma C, Yi B, Jing W, Shen J. A male sterility-associated cytotoxic protein ORF288 in Brassica juncea causes aborted pollen development. J Exp Bot. 2012;63(3):1285–95.
Peng X, Wang K, Hu C, Zhu Y, Wang T, Yang J, Tong J, Li S, Zhu Y. The mitochondrial gene orfH79 plays a critical role in impairing both male gametophyte development and root growth in CMS-Honglian rice. BMC Plant Biol. 2010;10(1):125.
Grelon M, Budar F, Bonhomme S, Pelletier G. Ogura cytoplasmic male-sterility (CMS)-associated orf138 is translated into a mitochondrial membrane polypeptide in male-sterile Brassica cybrids. Mol Gen Genet. 1994;243(5):540.
Monéger F, Smart CJ, Leaver CJ. Nuclear restoration of cytoplasmic male sterility in sunflower is associated with the tissue-specific regulation of a novel mitochondrial gene. EMBO J. 1994;13(1):8–17.
Korth KL, Levings CS. Baculovirus expression of the maize mitochondrial protein URF13 confers insecticidal activity in cell cultures and larvae. Proc Natl Acad Sci U S A. 1993;90(8):3388–92.
Palmer JD. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19(1):325–54.
Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66(1):34–44.
Chang S, Yang T, Du T, Huang Y, Chen J, Yan J, He J, Guan R. Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica. BMC Genomics. 2011;12(1):497.
Jo YD, Choi Y, Kim DH, Kim BD, Kang BC. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genomics. 2014;15(1):561.
Hu Q, Hua W, Yin Y, Zhang X, Liu L, Shi J, Zhao Y, Qin L, Chen C, Wang H. Rapeseed research and production in China. Crop Journal. 2016;5(2):127–35.
Hu Q, Li YC, Mei DS, Fang XP, Lise NH, Sven BA. Establishment and identification of cytoplasmic male sterility in Brassica napus by intergeneric somatic hybridization (in Chinese with English abstract). Agric Sci China. 2003;2(12):1321–8.
Arimura S, Yanase S, Tsutsumi N, Koizuka N. The mitochondrial genome of an asymmetrically cell-fused rapeseed, Brassica napus, containing a radish-derived cytoplasmic male sterility-associated gene. Genes & Genetic Systems. 2018;93(4):143–8.
Li P, Kang L, Wang A, Cui C, Jiang L, Guo S, Ge X, Li Z. Development of a fertility restorer for inap CMS (Isatis indigotica) Brassica napus through genetic introgression of one alien addition. Front Plant Sci. 2019;10:257.
Wu Z, Hu K, Yan M, Song L, Wen J, Ma C, Shen J, Fu T, Yi B, Tu J. Mitochondrial genome and transcriptome analysis of five alloplasmic male-sterile lines in Brassica juncea. BMC Genomics. 2019;20(1):348.
Yamagishi H, Bhat SR. Cytoplasmic male sterility in Brassicaceae crops. Breed Sci. 2014;64(1):38.
Hu Q, Andersen S, Dixelius C, Hansen L. Production of fertile intergeneric somatic hybrids between Brassica napus and Sinapis arvensis for the enrichment of the rapeseed gene pool. Plant Cell Rep. 2002;21(2):147–52.
Yan X, Dong C, Yu J, Liu W, Jiang C, Liu J, Hu Q, Fang X, Wei W. Transcriptome profile analysis of young floral buds of fertile and sterile plants from the self-pollinated offspring of the hybrid between novel restorer line NR1 and Nsa CMS line in Brassica napus. BMC Genomics. 2013;14(1):26.
Liu J, Xiang R, Wang W, Mei D, Li Y, Mason AS, Li F, Hu Q. Cytological and molecular analysis of Nsa CMS in Brassica napus L. Euphytica. 2015;206(2):279–86.
Ding B, Hao M, Mei D, Zaman QU, Sang S, Wang H, Wang W, Li F, Cheng H, Hu Q. Transcriptome and hormone comparison of three cytoplasmic male sterile Systems in Brassica napus. Int J Mol Sci. 2018;19(12):4022.
Chen J, Guan R, Chang S, Du T, Zhang H, Xing H. Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS One. 2011;6(3):e17662.
Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31(20):5907–16.
Mackenzie S, Mcintosh L. Higher plant mitochondria. Plant Cell. 1999;11(4):571–86.
Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46(3):123–39.
Leino M, Teixeira R, Landgren M, Glimelius K. Brassica napus lines with rearranged Arabidopsis mitochondria display CMS and a range of developmental aberrations. Theor Appl Genet. 2003;106(7):1156–63.
Chen L, Guo X, Xie C, He L, Cai X, Tian L, Song B, Liu J. Nuclear and cytoplasmic genome components of Solanum tuberosum + S. chacoense somatic hybrids and three SSR alleles related to bacterial wilt resistance. Theor Appl Genet. 2013;126(7):1861–72.
Wang J, Jiang J, Li X, Li A, Zhang Y, Guan R, Wang Y. Complete sequence of heterogenous-composition mitochondrial genome ( Brassica napus ) and its exogenous source. BMC Genomics. 2012;13(1):675.
Belliard G, Pelletier G, Vedel F, Quetier F. Morphological characteristics and chloroplast DNA distribution in different cytoplasmic parasexual hybrids of Nicotiana tabacum. Mol Gen Genet. 1978;165(3):231–7.
Belliard G, Vedel F, Pelletier G. Mitochondrial recombination in cytoplasmic hybrids of Nicotiana tabacum by protoplast fusion. Nature. 1979;281(5730):401–3.
Noyszewski AK, Ghavami F, Alnemer LM, Soltani A, Gu YQ, Huo N, Meinhardt S, Kianian PM, Kianian SF. Accelerated evolution of the mitochondrial genome in an alloplasmic line of durum wheat. BMC Genomics. 2014;15(1):67.
Pelletier G, Primard C, Vedel F, Chetrit P, Remy R. Rousselle, Renard. Intergeneric cytoplasmic hybridization in Cruciferae by protoplast fusion. Mol Gen Genet. 1983;191(2):244–50.
Bhatnagar-Mathur P, Gupta R, Reddy PS, Reddy BP, Reddy DS, Sameerkumar CV, Saxena RK, Sharma KK. A novel mitochondrial orf147 causes cytoplasmic male sterility in pigeonpea by modulating aberrant anther dehiscence. Plant Mol Biol. 2018;97(1–2):1–17.
Xie H, Peng X, Qian M, Cai Y, Ding X, Chen Q, Cai Q, Zhu Y, Yan L, Cai Y. The chimeric mitochondrial gene orf182 causes non-pollen-type abortion in Dongxiang cytoplasmic male-sterile rice. Plant J. 2018;95(4):715–26.
Igarashi K, Kazama T, Motomura K, Toriyama K. Whole genomic sequencing of RT98 mitochondria derived from Oryza rufipogon and northern blot analysis to uncover a cytoplasmic male sterility-associated gene. Plant Cell Physiol. 2013;54(2):237–43.
Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T. Gene organization deduced from the complete sequence of livewort Marchantia polymorpha mitochondrial DNA primitive form of plant mitochondrial genome. J Mol Biol. 1992;223(1):1–7.
Gray MW, Lang BL, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn TG, et al. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26(4):865–78.
Huang WC, Hu J, Zhu RS, Li SQ, Wang K, Yu JH, Li YS, Zhang ZH, Zhu YG. Research and development of the HL-type cytoplasmic male sterility rice (in Chinese). Science China Life Science. 2012;42:689–98.
Luo D, Xu H, Liu Z, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet. 2013;45(5):573.
Warmke HE, Lee SL. Pollen abortion in T cytoplasmic male-sterile corn (Zea mays): a suggested mechanism. Science. 1978;200(4341):561–3.
Kim DH, Kang JG, Kim BD. Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol Biol. 2007;63(4):519–32.
Yamamoto MP, Kubo T, Mikami T. The 5-leader sequence of sugar beet mitochondrial atp6 encodes a novel polypeptide that is characteristic of Owen cytoplasmic male sterility. Mol Gen Genomics. 2005;273(4):342–9.
Wang K, Gao F, Ji Y, Liu Y, Dan Z, Yang P, Zhu Y, Li S. 2013. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. New Phytol. 2013;198(2):408–18.
Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18(3):676–87.
Rhoads DM, Levings CS III, Siedow JN. URF13, a ligand-gated, pore-forming receptor for T-toxin in the inner membrane of CMS-T mitochondria. J Bioenerg Biomembr. 1995;27(4):437–45.
Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS wed servers for the detection of tRNAs and snoRNAs. Nucleic acids research. 2005;33:(Web Server issue):686–689.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–5.
Darling AE, Mau B, Perna NT. rogressive Mauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS One. 2010;5(6):e11147.
Acknowledgements
We would like to thank professor Guan Rongzhan for assistance with guidance of organelle extraction. Sequencing services were provided by Personal Biotechnology Co., Ltd. in Shanghai, China.
Funding
This work was supported by the Natural Science Foundation of China (30871553), the Key program of Technological Innovation in Hubei Province (No. 2016ABA084), the Science and Technology Innovation Project of Chinese Academy of Agricultural Sciences (Group No. 118), the Hubei Agricultural Science and Technology Innovation Center (201620000001048).
The funders had no role in the designing and conducting of this study and collection, analysis, and interpretation of data and writing the manuscript.
Author information
Authors and Affiliations
Contributions
SSF, CHT and HQ designed the experiment. SSF performed most experiments and bioinformatics analyses, and drafted the manuscript. LJ and HMY participated in organelle genome DNA preparation. QUZ, CHT and HQ revised the manuscript. ZHY helped on analysis the protein-coding gene analysis. MDS, FL and WH prepared plant material, HQ supervised the study. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Comparative analysis of Nsa CMS, Zhongshuang 4, and S. arvensis chloroplast (cp) genomes. (A) Comparison between Nsa CMS cp genome (vertical axis) and Zhongshuang 4 cp genome (horizontal axis) indicated that the nucleotide sequences of the syntenic region are well conserved. (B) Alignment of Nsa CMS cp genome (vertical axis) and S. arvensis cp genome (horizontal axis). Apart from SNPs, they were consistent and no rearrangement was found.
Additional file 2.
Primers for verification of mitochondrial genome recombination.
Additional file 3.
Sequence information of genes involved in phylogenetic tree construction.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sang, SF., Mei, DS., Liu, J. et al. Organelle genome composition and candidate gene identification for Nsa cytoplasmic male sterility in Brassica napus. BMC Genomics 20, 813 (2019). https://doi.org/10.1186/s12864-019-6187-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6187-y