Abstract
Background
The plant-specific gibberellic acid stimulated Arabidopsis (GASA) gene family is critical for plant development. However, little is known about these genes, particularly in fruit tree species.
Results
We identified 15 putative Arabidopsis thaliana GASA (AtGASA) and 26 apple GASA (MdGASA) genes. The identified genes were then characterized (e.g., chromosomal location, structure, and evolutionary relationships). All of the identified A. thaliana and apple GASA proteins included a conserved GASA domain and exhibited similar characteristics. Specifically, the MdGASA expression levels in various tissues and organs were analyzed based on an online gene expression profile and by qRT-PCR. These genes were more highly expressed in the leaves, buds, and fruits compared with the seeds, roots, and seedlings. MdGASA genes were also responsive to gibberellic acid (GA3) and abscisic acid treatments. Additionally, transcriptome sequencing results revealed seven potential flowering-related MdGASA genes. We analyzed the expression levels of these genes in response to flowering-related treatments (GA3, 6-benzylaminopurine, and sugar) and in apple varieties that differed in terms of flowering (‘Nagafu No. 2’ and ‘Yanfu No. 6’) during the flower induction period. These candidate MdGASA genes exhibited diverse expression patterns. The expression levels of six MdGASA genes were inhibited by GA3, while the expression of one gene was up-regulated. Additionally, there were expression-level differences induced by the 6-benzylaminopurine and sugar treatments during the flower induction stage, as well as in the different flowering varieties.
Conclusion
This study represents the first comprehensive investigation of the A. thaliana and apple GASA gene families. Our data may provide useful clues for future studies and may support the hypotheses regarding the role of GASA proteins during the flower induction stage in fruit tree species.
Similar content being viewed by others
Background
There has recently been an increase in the number of studies regarding low-molecular weight peptides. Gibberellic acid stimulated Arabidopsis (GASA), which is a cysteine-rich peptide, is an example of a low-molecular weight peptide important for plant growth and development.
The GAST1 gene, which was first identified in the gib1 tomato mutant, belongs to the first identified GASA gene family [1]. Many GASA homologs have recently been identified in Arabidopsis thaliana, tomato, rice, potato, wheat, and Petunia hybrida [2,3,4,5,6]. The GASA proteins typically consist of 80–270 amino acids, but there are exceptions (e.g., AtGASA14). The GASA genes encode small proteins with the following three domains [7]: (1) an N-terminal signal peptide with 18–29 amino acids, (2) a highly variable hydrophilic region with 7–31 polar amino acid residues, and (3) a C-terminal GASA domain consisting of 60 amino acids, typically including 12 cysteine residues [8, 9]. Additionally, previous studies revealed that peptides with a mutated or missing GASA domain are non-functional [10, 11].
Clarifying the subcellular localization of proteins may provide important clues regarding function. Most identified GASA proteins are reportedly localized in the cell wall or apoplast, with the signal peptide serving a critical function related to protein trafficking and localization [3, 4, 12]. For example, AtGASA4 and AtGASA6 are normally present at the cell periphery, but localize in the nucleus if lacking the signal peptide [9]. Additionally, two other GASA proteins, GIP2 and GIP5, accumulate in the cell well in petunia [3]. Meanwhile, an earlier study involving recombinant OsGASR-GFP proteins revealed that OsGASR1 and OsGASR2 localize to the apoplast or cell wall [4]. What’s more, OsGSR1 was detected in the plasma membrane, cytoplasm, and nucleus [13].
In plants, some GASA proteins have been functionally characterized and reportedly affect several processes during growth and development, including defense responses against pathogens and fungi, and stress resistance [10, 14,15,15,17]. Other studies have confirmed that GASA proteins influence hormone-related processes such as seed germination, floral development, stem elongation, root development, and signal transduction [3, 10, 12, 13, 18, 19]. For example, OsGSR1 can interact with DIM/DWF1, which is a brassinosteroid synthetase, to influence brassinosteroid signaling in rice [13]. Additionally, Most GASA family members are involved in gibberellic acid (GA3) signaling [4, 18], while some are also associated with abscisic acid (ABA), naphthaleneacetic acid, and indole-3-acetic acid signaling. For example, AtGASA2/3, AtGASA5, and AtGASA14 have been linked to ABA signaling [9]. GASA family members may exhibit opposing functions. Researchers have confirmed that AtGASA4 promotes flowering, while AtGASA5 induces the opposite effect [7, 12, 20]. Additionally, in Gerbera hybrida, the proteins encoded by GEG and PRGL, which are two GASA homologs, have different functions regarding floral development. Earlier studies demonstrated that GEG inhibits petal elongation, while PRGL induces petal elongation [21, 22]. Among their biological activities, their effect on flowering is one of the most prominent. Plants over-expressing AtGASA5 reportedly exhibit a late-flowering phenotype as well as down-regulated expression of FT and LFY, but up-regulated FLC expression [12]. In contrast, AtGASA4 expression promotes floral development [20]. Additionally, overexpressing FaGAST2 showed delayed growth in strawberry [23]. However, very little is known about GASA genes in woody plants as well as apple.
In contrast to the GASA genes in the model plant, A. thaliana, as well as in other plant species, which have been studied, little is known about the GASA genes in perennial woody species. Apple, as an important fruit tree species, is widely cultivated in temperate regions. The induction of flowering is an important consideration for apple producers and breeders. Most widely grown apple cultivars have a long juvenile period and exhibit poor flower bud development, which is problematic for the apple industry [24, 25]. Flower induction in apple trees is mediated by a complex biological process involving several important gene families, including SPL, MADs-box, and IDD [24, 26, 27]. Thus, identifying apple GASA family members and characterizing their potential roles will undoubtedly be useful. The sequenced apple genome [28] enables a whole-genome search for GASA genes as well as candidate genes responsible for inducing flower bud development. In this study, we first identified the GASA genes in the A. thaliana and apple genomes. We then analyzed the gene structures and classifications as well as phylogenetic relationships to characterize the apple GASA genes. Furthermore, an analysis of tissue-specific and flowering-related gene expression revealed candidate GASA genes associated with flower induction. To our best understanding, this study represents the first comprehensive analysis of A. thaliana and apple GASA genes. Our data may serve as a valuable resource for future studies of GASA genes related to flowering in apple as well as in other fruit tree species.
Results
Genome-wide identification of Arabidopsis thaliana and Malus domestica GASA genes
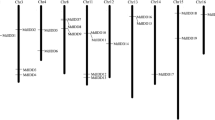
Thirteen GASA genes were previously identified in the A. thaliana TAIR7.0 genome [20]. Another two AtGASA genes (AT3g10185 and AT1g10588) were then verified and replenished in the Arabidopsis genome. These two added new genes were named AtGASA14 and AtGASA15. And these 15 AtGASA genes all shared conserved 12 cysteines (Additional file 1). To identify apple GASA genes, a BLASTP search of the apple genome was completed with the 15 AtGASA protein sequences used as queries. Furthermore, the 26 putative apple GASA genes were manually checked and confirmed using the conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Table 1). They were named according to their chromosomal locations (MdGASA1–26) (Fig. 1). The 26 MdGASA genes were located in 11 chromosomes in the apple genome. The chromosomes 9 and 17 contain most of the genes with 6 genes each, while the chromosomes 4, 5, 13, 14, 15, and 16 contain 1 copy each. Chromosome 8 and 12 contained three genes (Fig. 1).
The full MdGASA protein sequences were aligned to examine whether a GASA domain was present (Fig. 2b). All of the putative MdGASA proteins shared a conserved GASA domain, except for MdGASA24 and MdGASA10, whose GASA domains were mutated by the insertion of several amino acids.
Alignment of the GASA domain from MdGASA proteins. a Analysis of GASA protein structures, red column represented their conserved twelve cysteines. b Multiple alignments of the MdGASA protein sequences. Their conserved GASA domains were indicated. c Sequence logo analysis of the conserved GASA domains. Each stack represented their amino acids
Gene characterization and structure analysis of MdGASA
Protein characteristics, including molecular weight, isoelectric point, instability index, grand average of hydropathicity (GRAVY), major amino acid content, and aliphatic index, were analyzed with the ExPASy program. The molecular weight of the analyzed GASA proteins ranged from 9.44 (AtGASA8) to 33.97 (MdGASA24). Additionally, the molecular weight of most of the GASA proteins was less than 13 (i.e., low-molecular weight peptides). Moreover, the isoelectric point ranged from 7.41(AtGASA1) to 10.14 (MdGASA24) (Table 2). Most of the GASA proteins were observed to be unstable, with instability index values greater than 40. The exceptions were AtGASA5, AtGASA6, AtGASA9, AtGASA10, AtGASA12, MdGASA1, MdGASA4, and MdGASA7. According to the GRAVY values, the GASA proteins were hydrophilic, except for AtGASA6, AtGASA610, AtGASA11, AtGASA12, MdGASA5, MdGASA6, MdGASA14, MdGASA15, and MdGASA16. Meanwhile, the aliphatic index values ranged from 84.65 (AtGASA12) to 41.67 (MdGASA11). In terms of amino acid content, Cys, Lys, and Leu were the predominant residues, while Ser, Asp, Val, Pro, and Thr were also detected among the A. thaliana and apple GASA proteins. We also analyzed the transmembrane helices of all MdGASA proteins. At least one transmembrane segment was detected for MdGASA3, MdGASA6, MdGASA11, MdGASA12, MdGASA13, MdGASA18, MdGASA20, MdGASA25, and MdGASA26 (Additional file 2). The predicted protein structures for all MdGASA proteins (Fig. 3, Additional file 3) revealed the presence of α helices, β sheets, extended strands, and random coils. Of these structures, random coils were the most abundant, while β sheets were the least common. Moreover, the random coils were larger than the α helices.
Exon-intron structures were generated based on the annotated apple genome using the Gene Structure Display Server program. The MdGASA genes within each group shared conserved genetic structures (Fig. 4b). For example, the Group 2 genes, which included MdGASA4, MdGASA19, MdGASA7, MdGASA1, and MdGASA6, all contained one intron and two exons. Meanwhile, MdGASA2, MdGASA18, MdGASA11, and MdGASA25 were highly conserved and comprised four exons and three introns. Similar results were observed for the Group 3 genes (MdGASA14, MdGASA16, MdGASA17, and MdGASA20), which consisted of three exons and two introns. Furthermore, we detected four conserved protein motifs among the MdGASA proteins (Fig. 4c, Additional file 4). Members of Groups 1 and 2 carried motifs 1 and 2 except MdGASA3. G1 and G2 all shared motif 1 and motif 2. However, in Group 3, MdGASA15 contained only motif 1, while, MdGASA8, MdGASA21, MdGASA22, and MdGASA23 consisted of all four motifs.
Analysis of synteny and evolutionary relationships among GASA genes
To clarify the evolutionary relationships among GASA genes, we constructed a phylogenetic tree based on the A. thaliana and apple GASA protein sequences. According to the phylogenetic tree (Fig. 5), the A. thaliana and apple GASA genes were classified into three groups, with Groups 1, 2, and 3 consisting of 13, 9, and 19 GASA genes, respectively. Eight apple genes (MdGASA2, MdGASA18, MdGASA11, MdGASA25, MdGASA13, MdGASA26, MdGASA3, and MdGASA12) were clustered in Group 1, while five genes (MdGASA4, MdGASA9, MdGASA7, MdGASA1, and MdGASA6) were clustered in Group 2 and 13 genes (MdGASA10, MdGASA21, MdGASA22, MdGASA23, MdGASA8, MdGASA9, MdGASA24, MdGASA5, MdGASA15, MdGASA14, MdGASA16, MdGASA17, and MdGASA20) were clustered in Group 3.
To characterize the expansion patterns of the MdGASA genes, a diagram prepared with the Circos program was used to examine the duplicated blocks in the apple genome. Finally, two pairs of MdGASA genes (MdGASA7–MdGASA19 and MdGASA9–MdGASA22) were distributed among four chromosomes (Fig. 6a). Additionally, these duplicated MdGASA genes were from chromosomes with many genes, including chromosomes 8, 9, and 17. The exception was MdGASA19, which was located on chromosome 15.
Analysis of evolutionary relationships among GASA gene family members. Relative positive positions were depicted according to the apple chromosomes, colored lines were syntenic regions of the apple genome. (b) Synteny analysis of GASA genes between Arabidopsis and apple, relative positive positions were depicted according to the apple and Arabidopsis chromosomes, colored lines were syntenic regions of apple and Arabidopsis genome
Additional diagrams were prepared using the Circos program to evaluate the evolutionary relationships among A. thaliana and apple GASA genes (Fig. 6b). Two A. thaliana–apple GASA gene pairs (AtGASA13–MdGASA9 and AtGASA4–MdGASA22) were identified from three chromosomes. These two paired genes were detected in syntenic genomic regions. Additionally, both of the duplicated AtGASA genes were located on chromosome 5, while the two duplicated MdGASA genes were located on chromosomes 9 and 17.
MdGASA expression patterns in different tissues or organs
While A. thaliana GASA genes have been relatively well characterized, little is known about the expression of apple GASA genes. Thus, we systematically investigated their expression patterns in different tissues or organs using an online ArrayExpress database (E-GEOD- GSE42873) and a quantitative real-time polymerase chain reaction (qRT-PCR). Seven tissues or organs (leaves, flowers, fruits, seeds, stems, roots, seedlings) from 10 apple varieties (‘M67’, ‘M74’, ‘M20’, ‘M14’, ‘M9’, ‘M74’, ‘GD’, ‘X8877’, and two hybrids) were analyzed. The 26 candidate MdGASA genes exhibited diverse expression patterns among the various tissues (Fig. 7). All of the MdGASA genes were more highly expressed in the flowers, fruits, and leaves than in the stems and seedlings (Fig. 7). Moreover, MdGASA3, MdGASA14, and MdGASA20 were hardly expressed in all tissues and varieties, while MdGASA16 was highly expressed only in ‘M74’ flowers and ‘M20’ fruits.
To further analyze the potential MdGASA functions related to apple growth and development, different tissues (stems, leaves, flowers, fruits, and buds) were collected from ‘Nagafu No. 2’ trees. Because the MdGASA genes were extremely short and included a highly conserved GASA domain, 17 primer pairs were only designed to analyze expression levels [24, 26]. We observed diverse MdGASA expression patterns in ‘Nagafu No. 2’ tissues (Fig. 8). For example, MdGASA3 and MdGASA13/26 were highly expressed in fruits, while MdGASA1/6/7/19, MdGASA5, MdGASA15, and MdGASA24 expression levels were high in leaves. In contrast, MdGASA2, MdGASA4, MdGASA11/25, MdGASA14, MdGASA17/20, MdGASA18, and MdGASA21/22/23 were highly expressed in buds.
Effect of GA3 and ABA treatments on the expression of MdGASA genes in apple leaves
To elucidate the effects of phytohormones on MdGASA expression, trees were treated with GA3 and ABA, and leaves were collected at 3, 6, 12, and 24 h after treatments. As shown in Additional file 5, the MdGASA genes were responsive to exogenously applied GA3 and ABA, with the exception of MdGASA4, MdGASA17/20, and MdGASA24. MdGASA11/25, MdGASA15, and MdGASA21/22/23, were down-regulated by GA3 during the sampling period or at most time points. Meanwhile, MdGASA13/26 expression was inhibited at all time points. Furthermore, the largest increase in GA3-induced MdGASA expression was observed for MdGASA18 at 3 h after treatment.
MdGASA expression patterns in response to GA3 and 6-benzylaminopurine treatments during the flower induction period
To assess the potential effects of MdGASA expression induced by GA3 or 6-benzylaminopurine (6-BA) on apple flower induction, the transcript levels of seven candidate genes were investigated (Additional file 6) [29]. As shown in Fig. 9a, MdGASA1/6/7/19 expression was up-regulated by GA3 at all time points. Additionally, MdGASA5 expression was initially up-regulated, but was subsequently down-regulated during the flower induction period. The transcript levels for the other candidate genes were down-regulated by exogenously applied GA3. In contrast, MdGASA expression patterns varied over time in response to 6-BA (Fig. 9b). For example, MdGASA1/6/7/19 expression was inhibited only at 40, 50, and 70 days after full bloom (DAFB). In contrast, MdGASA5 expression was up-regulated by 6-BA except at 40 DAFB. Moreover, MdGASA8/9 expression was inhibited only at 30 and 70 DAFB, while MdGASA13/26 expression was inhibited at 40, 50, and 70 DAFB. Furthermore, MdGASA21/22/23 expression was up-regulated except at 30 and 70 DAFB, while MdGASA11/25 and MdGASA17/20 expression was inhibited by 6-BA at all time points.
Analysis of flowering-related MdGASA expression levels in response to GA3 (a) and 6-BA (b) treatments. Samples were collected at 30, 40, 50, 60, 70, and 80 days after full bloom (DAFB), with water used as a control. Each value represents the mean ± standard error of three replicates. Means followed by small letters are significantly different at the 0.05 level
MdGASA expression patterns in response to sugar treatments and in different flowering varieties during the flower induction period
We also investigated MdGASA expression patterns following sugar treatments during the apple flower induction period. The MdGASA1/6/7/19 transcription levels fluctuated, and were inhibited at 30, 50, and 70 DAFB, while MdGASA5 expression was inhibited except at 30 and 40 DAFB. Additionally, MdGASA8/9 expression was down-regulated except at 50 DAFB. Meanwhile, MdGASA13/26 expression levels increased only at 30 and 60 DAFB. Similarly, MdGASA11/25 expression was down-regulated except at 30 and 60 DAFB. Expression-level differences were also detected between MdGASA17/20 and MdGASA21/22/23 (Fig. 10a). The considerable increases or decreases in MdGASA expression levels in response to sugar treatments implied these genes may be associated with sugar signaling pathways during the flower induction period.
Analysis of flowering-related MdGASA expression levels in response to sugar (a) and in different apple varieties (‘Nagafu No. 2’ and ‘Yanfu No. 6’) (b). Samples were collected at 30, 40, 50, 60, 70, and 80 days after full bloom (DAFB), with water used as a control. Each value represents the mean ± standard error of three replicates. Means followed by small letters are significantly different at the 0.05 level
We further analyzed the MdGASA expression levels in apple varieties that differed in terms of flowering (‘Nagafu No. 2’ and ‘Yanfu No. 6’) (Fig. 10b). MdGASA1/6/7/19 expression was down-regulated in ‘Yanfu No. 6’ at the three early sampling dates, but was then up-regulated. Moreover, the MdGASA5 expression level was lower in ‘Yanfu No. 6’ except at 40 DAFB, while MdGASA8/9 expression was higher in ‘Yanfu No. 6’ except at 70 DAFB. The expression levels of MdGASA13/26 and MdGASA11/25 were higher in ‘Yanfu No. 6’ only at 60 and 70 DAFB. The transcription level of MdGASA17/20 was lower in ‘Yanfu No. 6’ only at the first three time points, while MdGASA21/22/23 expression was higher in ‘Yanfu No. 6’ only at the final three time points.
Analysis of the cis-elements in the MdGASA promoters
To investigate the regulatory mechanisms of MdGASA genes, a 1.5-kb promoter region upstream of the start codon (ATG) was isolated based on the apple genome sequence (Fig. 11) and analyzed to identify potential cis-elements [24]. Several stress-related cis-elements were detected in the promoters of the 26 candidate MdGASA genes. Moreover, meristem-related cis-elements were also identified in the MdGASA1, MdGASA11, MdGASA13, and MdGASA15 promoters. Additional hormone-related cis-elements were detected in various MdGASA genes. These identified motifs indicated MdGASA genes might be regulated by cis-elements within the corresponding promoters.
Discussion
The low-molecular weight GASA proteins influence various biological processes and are important for plant growth and development. To our best understanding, a systematic identification of GASA genes has only been reported for Solanum tuberosum [5]. Little is known about the corresponding apple genes. In this study, we compiled an improved list of candidate A. thaliana GASA genes based on a published study [20]. We also identified the candidate apple GASA genes, and subsequently characterized the genes in terms of phylogenetic relationships, structure, synteny, and tissue-specific expression patterns. Finally, an analysis of MdGASA expression levels in response to various flowering-related factors indicated these genes may affect apple flower induction. This study represents the first comprehensive investigation of the A. thaliana and apple GASA gene families, and the resulting data will undoubtedly be useful for future analyses, including investigations on the potential roles for GASA proteins during hormone- or sugar-mediated flower induction in fruit trees.
Genome-wide identification and characterization of GASA genes in Arabidopsis and apple
We identified 15 and 26 GASA genes in the A. thaliana and apple genomes, respectively. The fact we detected more AtGASA genes than a previous study [20] may have been because we used an updated TAIR database. Additionally, the number of MdGASA genes was greater than the number of A. thaliana and potato GASA genes [5]. A subsequent analysis of the identified AtGASA and MdGASA genes confirmed they all encoded a conserved GASA domain containing 12 cysteine residues (Fig. 1, Additional file 1) [7,8,9].
We compared the A. thaliana and apple GASA proteins in terms of several characteristics, including length, molecular weight, isoelectric point, instability index, GRAVY, amino acid content, and aliphatic index. Interestingly, all of the identified GASA genes were shorter with a lower molecular weight than the members of other known flowering-related gene families (i.e., SPL, MADS-box, and IDD) [24, 26, 27, 30]. This observation is consistent with the fact GASA genes encode low-molecular weight proteins [2, 5, 31]. Additionally, the amino acid contents of the identified A. thaliana and apple GASA proteins were very similar (Table 2). The abundance of Cys residues among the GASA proteins was likely due to the highly conserved 12 cysteines (Fig. 1, Additional file 1). However, we observed differences in the instability index and GRAVY values between the AtGASA and MdGASA proteins, which may be associated with the variability in the intermediate regions (i.e., 7–31 polar amino acids) [5]. Furthermore, all of the identified MdGASA proteins shared similar structures and motifs (e.g., motif 1) (Fig. 4b). Additionally, genes from the same group shared similar exon-intron structures, indicating that the evolution of the GASA domain was associated with genetic structures. However, we detected some structural differences among the GASA genes, and the gain or loss of exons or introns may have been a consequence of chromosomal rearrangements [32, 33].
All of the identified AtGASA genes were equally distributed on five A. thaliana chromosomes (Fig. 5b), while the 26 MdGASA genes were located on only 11 of 17 apple chromosomes. Six apple chromosomes (chromosomes 1, 2, 6, 7, 10 and 11) did not contain any copy of the MdGASA genes. A similarly uneven distribution of GASA genes was reported for potato [5].
Evolutionary and syntenic relationships among GASA genes
Phylogenetic and syntenic relationships among GASA genes were analyzed. First, we constructed a phylogenetic tree based on the A. thaliana and apple GASA protein sequences (Fig. 3). The 41 GASA protein sequences were clustered into three groups, which were consistent with the phylogenetic classification of potato homologs [5]. The GASA genes were nearly equally distributed among the three groups. However, MdGASA10 did not cluster with the other GASA genes according to the phylogenetic tree. This may have been because of the insertion of several amino acids in the GASA domain (Fig. 2). Additionally, the AtGASA and MdGASA genes were closely clustered, possibly because A. thaliana and apple are both dicotyledonous species that may have a common ancestor.
A previous study revealed that tandem, segmental, and whole genome duplications are important for the evolution of species [32]. To date, some apple gene duplications have been characterized (e.g., SPL, IDD, and bZIP genes) [24, 26, 34]. In the current study, we investigated the duplication of MdGASA genes. Two gene pairs (MdGASA7–MdGASA19 and MdGASA9–MdGASA22) were likely the result of segmental duplications (Fig. 5a). Additionally, these duplicated genes clustered together in the same phylogenetic group and their expression patterns were suggested to be relatively stable [35]. An earlier study concluded that a recent genome duplication event promoted the expansion of the apple genome, leading to an increase in the number of chromosomes from nine to 17 [28]. Thus, genome duplications have played an important role in the expansion of MdGASA genes. Moreover, this gene duplication and expansion may have contributed to an increase in the diversity of MdGASA genes regarding quality, structure and function.
Genomic comparisons in which a newly identified gene is compared with orthologous genes from well-characterized plant species are useful for generating information and providing valuable clues regarding gene structures and functions [36,37,38]. In the current study, we developed a method to analyze syntenic relationships between the apple genome and the well-characterized A. thaliana genome to elucidate evolutionary relationships and possible roles of MdGASA genes (Fig. 5b). Although potential roles for genes can be ascribed based on the known functions of several well-characterized homologs, further research will be needed to confirm the putative protein functions.
MdGASA gene expression profiles and potential functions
We investigated the expression patterns of MdGASA genes in different apple varieties and tissues based on the ArrayExpress database and qRT-PCR. The Group 2 genes (MdGASA4, MdGASA19, MdGASA7, MdGASA1 and MdGASA6) shared similar expression patterns, which was consistent with the similarities in their gene structures (Fig. 4a). In contrast, the genes from Groups 1 and 3 exhibited diverse expression patterns among various tissues. Interestingly, the expression levels of almost all of the identified MdGASA genes were down-regulated in seeds or seedlings (Fig. 6). Furthermore, MdGASA2, MdGASA4, MdGASA9, and MdGASA25 were most highly expressed in leaves, implying they may be important for leaf development [38, 39]. Meanwhile, MdGASA19, MdGASA21, MdGASA22, and MdGASA23 expression levels were highest in harvested fruits, suggesting their importance in developing fruits [23, 40]. Overall, the various expression patterns were consistent with the differences in gene chromosomal locations, characteristics, and structures.
We further analyzed the MdGASA expression patterns among different ‘Nagafu No. 2’ tissues (Fig. 7). Because of the similarities in the GASA genes and the fact their coding regions were very short, distinguishing between genes was difficult. Thus, only 17 primer pairs were designed for expression analyses. MdGASA genes (except MdGASA3 and MdGASA13/26) were highly expressed in the leaves, buds, and flowers, which suggested they play key roles in the development of these tissues [7, 20, 21]. Additionally, in floral biology research, the leaves and buds have traditionally been the important tissues used for analyses of flower induction [41, 42]. Additionally, MdGASA3 and MdGASA13/26 expression levels were highest in ‘Nagafu No. 2’ fruits, implying these genes have important effects related to fruit development [23, 40]. However, these roles will need to be verified in future studies.
The GASA proteins are important for regulating flower induction in various species such as A. thaliana [12, 20, 38], rice [4], strawberry [43], G. hybrida [21, 22], and P. hybrida [3]. However, little is known about their potential roles in regulating flower induction in apple. Consequently, we first investigated MdGASA expression patterns to assess whether they are associated with hormone- or sugar-mediated flower induction (Figs 8 and 9). Previous studies confirmed that hormones and sugars have important functions during the flower induction period, and that the application of exogenous GA3 decreases the flowering rates of apple trees, while sugar and 6-BA have the opposite effect [24, 44, 45]. We observed that our exogenous GA3 treatment almost inhibited the expression of the flowering-related MdGASA genes (MdGASA8, MdGASA13/26, MdGASA11/25, MdGASA17/20, and MdGASA21/22/23), which is consistent with the previously reported low flowering rate of GA3-treated trees [24, 45]. However, the MdGASA1/6/7/19 expression patterns differed from those of other flowering-related MdGASA genes in response to a GA3 treatment. The differences in expression patterns might be due to the redundant functions of GASA gene family members. For example, over-expressing GASA5 reportedly delays flowering, while over-expressing GASA6 leads to an early-flowering phenotype [12, 38]. Overall, our findings indicate that GASA genes are involved in GA-mediated flower induction.
Researchers have demonstrated that 6-BA positively affects apple flower induction [44, 46,47,48]. Several identified gene families are also involved in regulating flower induction in response to exogenous 6-BA treatments [24]. Sugar, as an energy source, is also important for initiating floral development activities [29, 42]. Sugar can be integrated by various flowering pathways to regulate flowering in apple [29]. However, whether GASA genes are associated with the relevant 6-BA or sugar signaling pathways has not been addressed. In the current study, we first analyzed the responses of the candidate flowering-related MdGASA genes to 6-BA or sugar treatments during the flower induction period (Figs 8b and 9a). The expression levels of most candidate MdGASA genes were up- or down-regulated by exogenous applications of 6-BA or sugar, indicating they may influence the regulation of flower induction by 6-BA or sugar. However, because of a relative lack of research on the effects of GASA proteins on 6-BA or sugar signaling, the hypothesis that GASA proteins influence the activities of 6-BA or sugar related to flower induction cannot be verified. Therefore, future research should focus on the potential relationships between MdGASA proteins and 6-BA or sugar signaling. Nevertheless, the results presented herein may be useful for summarizing the potential roles for MdGASA proteins in response to 6-BA and/or sugar.
The MdGASA genes exhibited different expression patterns in the ‘Nagafu No. 2’ and ‘Yanfu No. 6’ apple cultivars, which differ in terms of flowering. MdGASA1/6/7/9 and MdGASA11/25 exhibited the opposite expression profiles during the first three and final three analyzed time points of the floral induction stage. These observations were consistent with their expression patterns in response to exogenous GA3. Thus, these two apple genes likely have opposing functions regarding the regulation of flower induction, similar to AtGASA5 and AtGASA6 [12, 38]. However, this will need to be experimentally confirmed. Most of the GASA expression levels initially increased and then decreased in ‘Nagafu No. 2’ and ‘Yanfu No. 6’ trees. This expression pattern might be associated with differences in endogenous hormone levels, as ‘Nagafu No. 2’ trees reportedly require a greater abundance of hormones for growth [49]. To further investigate whether the expression of MdGASA genes is regulated by different hormones or signals, the cis-elements in the corresponding promoters were analyzed as previously described [24]. We observed that most of the GASA promoters had common hormone- and stress-related cis-elements, which may be involved in the complex regulatory mechanisms affecting gene expression (Fig. 10).
Conclusion
We identified 15 AtGASA genes and 26 MdGASA genes. All of the identified GASA genes included a conserved GASA domain, and exhibited similar physical and chemical characteristics. A phylogenetic analysis revealed that AtGASA and MdGASA genes can be classified into three groups. Additional analyses of synteny and gene duplications helped to elucidate the expansion and diversity of MdGASA genes. Moreover, an analysis of the spatiotemporal tissue-specific expression patterns indicated that most MdGASA genes were expressed more in the leaves, buds, and fruits than in the seeds and seedlings. Additionally, the expression levels of selected candidate flowering-related MdGASA genes were further analyzed in different varieties and in response to different treatments (i.e., GA3, 6-BA, and sugar). The resulting data indicated the MdGASA genes may help to regulate the induction of flowering in apple trees. Overall, our comprehensive genome-level investigation of A. thaliana and apple GASA genes provides basic relevant information for future studies. The data presented herein may help to support hypotheses regarding the involvement of GASA during the flower induction stage of fruit tree species.
Methods
Identification of Arabidopsis thaliana and apple GASA genes
AtGASA were identified from the Arabidopsis database (http://www.arabidopsis.org). Another two new GASA genes (AT3g10185 and AT1g10588) were replenished based on a previous study [20]. They were named AtGASA14 and AtGASA15.To identify apple GASA genes, we first used the 15 identified AtGASA protein sequences as queries to search the apple genome database (Malus domestica Genome v1.0, http://www.rosaceae.org/) (Additional file 7). The obtained sequences were then used as queries to search the conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). and to discard genes lacking a GASA domain. All non-redundant putative protein sequences were finally manually checked to confirm the presence of the GASA domain.
Chemical characterization, chromosome mapping and sequence alignments
The sequences of the candidate A. thaliana and apple GASA proteins were used to predict protein characteristics with the ExPASy program (http://web.expasy.org/protparam/). Additionally, the physical location of each MdGASA gene was determined according to the genome annotations. The genes were then mapped to specific chromosomes. The DNAMAN program was used to align the MdGASA and AtGASA protein sequences, while the WebLogo platform (http://weblogo.berkeley.edu/logo.cgi) was used to generate and analyze sequence logos. The TMHMM server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict the presence of transmembrane helices, while the predicted MdGASA protein structures were analyzed with the PHYRE server v2.0 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).
Analyses of phylogenetic relationships, gene structures and motifs, and promoters
A phylogenetic tree comprising all candidate A. thaliana and apple GASA genes was constructed using the neighbor-joining method of the MEGA 6.06 program. Default parameters were used with a bootstrap test involving 1000 replicates. The Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) was used to construct exon-intron structures. The gene structures were determined based on the coding sequences within the corresponding genomic sequence. The MEME platform (http://meme-suite.org/) was used to identify the conserved motifs in the MdGASA proteins [50] (default parameters with the maximum number of motifs set to 10). Furthermore, the 1.5-kb region upstream of the start codon of candidate MdGASA genes was examined for the presence of cis-elements. The PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to search for regulatory elements.
Plant materials and treatments
Two-year-old pot-cultivated ‘Nagafu No. 2’ apple trees grown on M.26 rootstocks underwent various treatments. Hormone treatments involved the application of 100 mΜ GA3 or 300 μM ABA to apple leaves, which were then collected at 0, 1, 3, 6, and 12 h after treatment.
Seventy-two uniformly growing 6-year-old ‘Fuji’/T337/Malus robusta Rehd. apple trees were randomly divided into four groups of 18 trees each. The groups were then treated with GA3, sucrose, 6-BA, or water (control). The trees were then grown at an experimental orchard in Yangling, China (108°04′ E, 34°16′ N). Each group was prepared as three blocks, with three replicates. The trees were analyzed from 30 to 80 DAFB in 2015. The GA3 treatment was completed using a slightly modified previously described method [45]. Briefly, 700 mg L−1 GA3 (Sigma, Deisenhofen, Germany) was sprayed once on a clear morning at 30 DAFB (May 9). Additionally, trees were sprayed with 300 mg L−1 6-BA (Sigma) on a clear morning at 30 DAFB (May 9). The sugar treatment involved spraying trees two times with 15,000 mg L−1 and 20,000 mg L−1 sucrose on clear mornings at 30 and 37 DAFB (May 9 and May 16). All treatments involved the whole tree and were applied with a low-pressure hand-wand sprayer. Terminal buds on the current-year spurs (< 5 cm), which were chosen according to previous studies [24, 25, 29], were collected at 30, 40, 50, 60, 70, and 80 DAFB and immediately frozen with liquid nitrogen and stored at −80 °C until used in gene expression analyses.
Buds from two apple varieties (‘Yanfu No. 6’ and ‘Nagafu No. 2’) were collected from 18 uniformly growing 6-year-old trees in 2015 at 30, 40, 50, 60, 70, and 80 DAFB. ‘Yanfu No. 6’ is a spontaneous mutant of ‘Nagafu No. 2’ that produces more flower buds. Moreover, ‘Yanfu No. 6’ trees produce a higher proportion of spurs, shorter internodes, bigger buds, and more flowers. Terminal buds on the current-year spurs (< 5 cm) were collected as described above.
Different organs were also collected for analyses of tissue-specific expression patterns. Flowers were collected at full bloom on April 9 in 2015. Additionally, stems were collected from new shoots with a diameter of 2–3 mm, while mature leaves were collected from the adjacent buds. Fruits with a diameter of 2–3 cm were also collected. All samples were immediately frozen in liquid nitrogen and stored at −80 °C until used in gene expression analyses.
RNA extraction and cDNA synthesis
Total RNA was extracted from plant tissue samples using a slightly modified cetyltrimethylammonium bromide (CTAB) method [51]. Briefly, 900 μL extraction buffer (2% CTAB, 2.5% PVP-40, 2 M NaCl, 100 mM Tris-HCl [pH 8.0], 25 mM EDTA [pH 8.0], and 2% β-mercaptoethanol) was pre-heated at 65 °C and added to 2-mL microcentrifuge tubes just before use. Bud samples (200 mg) stored at −80 °C were ground to a powder and then added to the extraction buffer in microcentrifuge tubes. After vigorously shaking and inverting each tube for 5 min and incubating at 65 °C for 30 min, an equal volume of chloroform:isoamyl alcohol (24:1, v/v) was added. The tube was vigorously shaken and inverted and then centrifuged at 12,000×g for 10 min at 4 °C. The supernatant was transferred to a new tube and re-extracted with an equal volume of chloroform:isoamyl alcohol (24:1, v/v). The supernatant was transferred to a new 2-mL tube, after which LiCl (3 M final concentration) was added. The mixture was incubated at −20 °C for 4 h, after which the RNA was selectively pelleted by LiCl after a centrifugation at 18,000×g for 20 min at 4 °C. The pellet was resuspended in 500 μL SSTE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% SDS, and 1 M NaCl) that had been pre-heated to 65 °C and an equal volume of chloroform:isoamyl alcohol. The mixture was then centrifuged at 12,000×g for 10 min at 4 °C. The supernatant was transferred to a new microcentrifuge tube, and the RNA was precipitated with 2.5 volumes of cold ethanol at −80 °C for at least 30 min. After a centrifugation at 12,000×g for 20 min at 4 °C, the pellets were washed with 70% ethanol and resuspended in diethylpyrocarbonate-treated water. Total RNA integrity was verified by 2% agarose gel electrophoresis. Additionally, first-strand cDNA was synthesized from 1 μg total RNA using a PrimeScript RT Reagent kit with gDNA Eraser (Takara Bio, Shiga, Japan).
Analysis of GASA expression
The expression patterns of candidate MdGASA genes were analyzed by qRT-PCR. All primer pairs were designed with the Primer 5.0 program (Additional file 8). However, because the coding sequences of the MdGASA genes were very short and highly conserved, we were unable to specifically amplify each gene. Thus, only 17 primer pairs were designed to analyze all 27 MdGASA genes. Two or three genes were amplified using previously designed primers [24, 26]. Consequently, MdGASA1, MdGASA6, MdGASA7, and MdGASA19 were amplified by the same primer pair. Additionally, MdGASA8 and MdGASA9, MdGASA11 and MdGASA25, MdGASA13 and MdGASA26, MdGASA17 and MdGASA20 were amplified by the same primer pairs, respectively. Finally, MdGASA21, MdGASA22, and MdGASA23 were also amplified by the same primers.
The qRT-PCR assay mix (20 μL) consisted of 2 μL cDNA (diluted 1:8), 10 μL 2× SYBR Premix ExTaq II (Takara Bio), 0.8 μL each primer (10 μM) (Table 1), and 6.4 μL distilled deionized H2O. Each qRT-PCR assay was completed on an iCycler iQ Real Time PCR Detection System (Bio-Rad) with the following program: 95 °C for 3 min; 40 cycles at 94 °C for 15 s, 62 °C for 20 s, and 72 °C for 20 s. The resulting fragments were immediately subjected to a melting-curve analysis to verify the presence of gene-specific PCR products. The melting-curve analysis was completed with the following program: 94 °C for 15 s, followed by a constant increase from 60 °C to 95 °C at a 2% ramping rate. The apple EF-1α gene (GenBank accession No. DQ341381) was used as an internal control and served as the standard for normalizing all mRNA levels. The 2−ΔΔCt method was used to calculate the relative template abundance in each PCR amplification mixture [52]. Three biological replicates were used for gene expression analysis [53].
Statistical analysis
Data underwent an analysis of variance and the means were compared by a t-test at the 5% level using the SPSS 11.5 software package (SPSS, Chicago, IL, USA). Figures were prepared using Origin 7.5 (Microcal Software Inc., Northampton, MA, USA).
Abbreviations
- 6-BA:
-
6-benzylaminopurine; ABA: abscisic acid
- DAFB:
-
Days after full blossom
- GA3 :
-
Gibberellic acid
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
References
Shi LF, Gast RT, Gopalraj M, Olszewski NE. Characterization of a shoot-specific, Ga3-regulated and Aba-regulated gene from tomato. Plant J. 1992;2(2):153–9.
Herzog M, Dorne AM, Grellet F. GASA, a gibberellin-regulated gene family from Arabidopsis Thaliana related to the tomato GAST1 gene. Plant Mol Biol. 1995;27(4):743–52.
Ben-Nissan G, Lee JY, Borohov A, Weiss D. GIP, a Petunia Hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J. 2004;37(2):229–38.
Furukawa T, Sakaguchi N, Shimada H. Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet Syst. 2006;81(3):171–80.
Nahirnak V, Rivarola M, de Urreta MG, Paniego N, Hopp HE, Almasia NI, Vazquez-Rovere C. Genome-wide analysis of the Snakin/GASA gene family in Solanum Tuberosum cv. Kennebec. Am J Potato Res. 2016;93(2):172–88.
Zhang LY, Geng XL, Zhang HY, Zhou CL, Zhao AJ, Wang F, Zhao Y, Tian XJ, Hu ZR, Xin MM, et al. Isolation and characterization of heat-responsive gene TaGASR1 from wheat (Triticum Aestivum L.). J Plant Biol. 2017;60(1):57–65.
Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis Thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol. 1998;36(6):871–83.
Silverstein KA, Moskal WA Jr, Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007;51(2):262–80.
Zhang SC, Wang XJ. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chinese Sci Bull. 2008;53(24):3839–46.
Rubinovich L, Weiss D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010;64(6):1018–27.
Sun S, Wang H, Yu H, Zhong C, Zhang X, Peng J, Wang X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J Exp Bot. 2013;64(6):1637–47.
Zhang S, Yang C, Peng J, Sun S, Wang X. GASA5, a regulator of flowering time and stem growth in Arabidopsis Thaliana. Plant Mol Biol. 2009;69(6):745–59.
Wang L, Wang Z, Xu YY, Joo SH, Kim SK, Xue Z, Xu ZH, Wang ZY, Chong K. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009;57(3):498–510.
Bindschedler LV, Whitelegge JP, Millar DJ, Bolwell GP. A two component chitin-binding protein from French bean - association of a proline-rich protein with a cysteine-rich polypeptide. FEBS Lett. 2006;580(6):1541–6.
Mao ZC, Zheng JY, Wang YS, Chen GH, Yang YH, Feng DX, Xie BY. The new CaSn gene belonging to the snakin family induces resistance against root-knot nematode infection in pepper. Phytoparasitica. 2011;39(2):151–64.
Balaji V, Smart CD. Over-expression of snakin-2 and extensin-like protein genes restricts pathogen invasiveness and enhances tolerance to Clavibacter michiganensis subsp michiganensis in transgenic tomato (Solanum Lycopersicum). Transgenic Res. 2012;21(1):23–37.
Wigoda N, Ben-Nissan G, Granot D, Schwartz A, Weiss D. The gibberellin-induced, cysteine-rich protein GIP2 from Petunia Hybrida exhibits in planta antioxidant activity. Plant J. 2006;48(5):796–805.
Li KL, Bai X, Li Y, Cai H, Ji W, Tang LL, Wen YD, Zhu YM. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J Plant Physiol. 2011;168(18):2153–60.
Zhang SC, Wang XJ. Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J Plant Physiol. 2011;168(17):2093–101.
Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant & cell physiology. 2007;48(3):471–83.
Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, Elomaa P, Teeri TH. GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera Hybrida. Plant Cell. 1999;11(6):1093–104.
Peng JZ, Lai LJ, Wang XJ. Temporal and spatial expression analysis of PRGL in Gerbera Hybrida. Mol Biol Rep. 2010;37(7):3311–7.
Moyano-Canete E, Bellido ML, Garcia-Caparros N, Medina-Puche L, Amil-Ruiz F, Gonzalez-Reyes JA, Caballero JL, Munoz-Blanco J, Blanco-Portales R. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013;54(2):218–36.
Fan S, Zhang D, Xing L, Qi S, Du L, Wu H, Shao H, Li Y, Ma J, Han M: Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus. Molecular genetics and genomics. MGG. 2017;292:755–71.
Xing LB, Zhang D, Zhao CP, Li YM, Ma JJ, An N, Han MY. Shoot bending promotes flower bud formation by miRNA-mediated regulation in apple (Malus Domestica Borkh.). Plant Biotechnol J. 2016;14(2):749–70.
Li J, Hou H, Li X, Xiang J, Yin X, Gao H, Zheng Y, Bassett CL, Wang X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.). Plant Physiol Biochem. 2013;70:100–14.
Kumar G, Arya P, Gupta K, Randhawa V, Acharya V, Singh AK: Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malus x domestica). Sci Rep-Uk. 2016;6:20695.
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet. 2010;42(10):833.
Xing LB, Zhang D, Li YM, Shen YW, Zhao CP, Ma JJ, An N, Han MY. Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (Malus Domestica Borkh.). Plant Cell Physiol. 2015;56(10):2052–68.
Tian Y, Dong QL, Ji ZR, Chi FM, Cong PH, Zhou ZS. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene. 2015;555(2):277–90.
Berrocal-Lobo M, Segura A, Moreno M, Lopez G, Garcia-Olmedo F, Molina A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002;128(3):951–61.
Xu GX, Guo CC, Shan HY, Kong HZ. Divergence of duplicate genes in exon-intron structure. P Natl Acad Sci USA. 2012;109(4):1187–92.
Guo RR, Xu XZ, Carole B, Li XQ, Gao M, Zheng Y, Wang XP. Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape. BMC Genomics. 2013;14
Zhao J, Guo RR, Guo CL, Hou HM, Wang XP, Gao H. Evolutionary and expression analyses of the apple basic Leucine zipper transcription factor family. Front Plant Sci. 2016;7
Zhang JZ. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18(6):292–8.
Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–38.
Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang HB, Wang XY, Bowers J, Paterson A, Lisch D, et al. Finding and comparing Syntenic regions among Arabidopsis and the Outgroups papaya, poplar, and grape: CoGe with Rosids. Plant Physiol. 2008;148(4):1772–81.
Qu J, Kang SG, Hah C, Jang JC. Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis Thaliana. Plant Sci. 2016;246:1–10.
Nahirnak V, Almasia NI, Fernandez PV, Hopp HE, Estevez JM, Carrari F, Vazquez-Rovere C. Potato Snakin-1 gene silencing affects cell division, primary metabolism, and Cell Wall composition. Plant Physiol. 2012;158(1):252–63.
Blanco-Portales R, Bellido ML, Garcia-Caparros N, Medina-Puche L, Caballero-Repullo JL, Gonzalez-Reyes JA, Munoz-Blanco J, Moyano E. The strawberry FaGAST2 gene determines receptacle cell size during fruit development and ripening. FEBS J. 2012;279:82.
Monerri C, Fortunato-Almeida A, Molina RV, Nebauer SG, Garcia-Luis A, Guardiola JL. Relation of carbohydrate reserves with the forthcoming crop, flower formation and photosynthetic rate, in the alternate bearing ‘Salustiana’ sweet orange (Citrus Sinensis L.). Sci Hortic-Amsterdam. 2011;129(1):71–8.
Fan S, Zhang D, Lei C, Chen HF, Xing LB, Ma JJ, Zhao CP, Han MY. Proteome analyses using iTRAQ labeling reveal critical mechanisms in alternate bearing Malus Prunifolia. J Proteome Res. 2016;15(10):3602–16.
de la Fuente JI, Amaya I, Castillejo C, Sanchez-Sevilla JF, Quesada MA, Botella MA, Valpuest V. The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J Exp Bot. 2006;57(10):2401–11.
Li YM, Zhang D, Xing LB, Zhang SW, Zhao CP, Han MY. Effect of exogenous 6-benzylaminopurine (6-BA) on branch type, floral induction and initiation, and related gene expression in ‘Fuji’ apple (Malus Domestica Borkh). Plant Growth Regul. 2016;79(1):65–70.
Zhang S, Zhang D, Fan S, Du L, Shen Y, Xing L, Li Y, Ma J, Han M. Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ apple (Malus Domestica Borkh.). Plant Physiol Biochem. 2016;107:178–86.
Ito A, Hayama H, Kaskimura Y, Yoshioka H. Effect of maleic hydrazide on endogenous cytokinin contents in lateral buds, and its possible role in flower bud formation on the Japanese pear shoot. Sci Hortic-Amsterdam. 2001;87(3):199–205.
Bernier G, Perilleux C. A physiological overview of the genetics of flowering time control. Plant Biotechnol J. 2005;3(1):3–16.
Krasniqi AL, Damerow L, Kunz A, Blanke MM. Quantifying key parameters as elicitors for alternate fruit bearing in cv. ‘Elstar’ apple trees. Plant Sci. 2013;212:10–4.
Song CH, Zhang D, Zheng LW, Zhang J, Zhang BJ, Luo WW, Li YM, Li GF, Ma JJ, Han MY. miRNA and Degradome sequencing reveal miRNA and their target genes that may mediate shoot growth in spur type mutant “Yanfu 6”. Front Plant Sci. 2017;8
Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue):W369–73.
Gambino G, Perrone I, Gribaudo I. A rapid and effective method for RNA extraction from different tissues of grapevine and other Woody plants. Phytochem Analysis. 2008;19(6):520–5.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25(4):402–8.
Fan S, Zhang D, Gao C, Zhao M, Wu H, Li Y, Shen Y, Han M. Identification, classification, and expression analysis of GRAS gene family in Malus Domestica. Front Physiol. 2017;8:253.
Acknowledgments
Not applicable.
Funding
China Apple Research System (CARS-28), National Natural Science Foundation of China (31672101), Science and Technology Innovative Engineering Project in Shaanxi province, China (2015NY114), China Postdoctoral Science Foundation (2014 M56806), Yangling Subsidiary Center Project of the National Apple Improvement Center and Collaborative Innovation of the Center for Shaanxi Fruit Industry Development (C000088). CARS-28, 2015NY114 and 31,672,101 were involved in sample collection. 2014 M56806 and C000088 were involved qPCR and manuscript revision.
Availability of data and material
The datasets supporting the conclusions of this article are included within the article and additional files.
Author information
Authors and Affiliations
Contributions
HM, FS and ZD conceived and designed the experiment. FS, GC, XM and ZL performed the experiment. FS, MMT, LY and MJ analyzed the data. HM and FS wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Malus domescica (‘Yanfu No. 6’ and ‘Nagafu No. 2’) were widely planted in China. They were not listed in the appendices I, II and III of the Convention on the Trade in Endangered Species of Wild Fauna and Flora (https://cites.org/eng/app/appendices.php). All the available materials used in our study were grown in the Apple Demonstration Nursery of Yangling Modern Agriculture Technology Park (Northwest A&F University). Samples collection complied with the institutional, national and international guidelines. This article did not contain any studies with human participants or animals performed byany of authors. No specific permits were required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Alignment of GASA domains from AtGASA proteins. (a) Multiple alignments of the AtGASA protein sequences and their conserved GASA domains, red column represented their conserved twelve cysteines. (b) Sequence logo analysis of the conserved AtGASA domains. Each stack represented their amino acids. (TIFF 5642 kb)
Additional file 2:
Transmembrane topology analysis of MdGASA proteins. Transmembrane helices of the MdGASA proteins were predicted with the TMHMM server v2.0. The red peaks indicate the predicted transmembrane helices. (TIFF 3853 kb)
Additional file 3:
Secondary structures of MdGASA protein. Their α helix, Extended strand, Random coil and β turn were analyzed. (DOCX 14 kb)
Additional file 4:
Motif sequence identified by MEME. Motif number was associated with fig. 4c. (TIFF 7247 kb)
Additional file 5:
Effects of GA3 and ABA on leaf MdGASA expression levels. Leaves were collected at 0, 1, 3, 6 and 12 h after each treatment. 100Μm GA3, and 300 μM ABA were sprayed in apple leaves. (TIFF 6945 kb)
Additional file 6:
MdGASA expression levels based on previous RNA sequencing data. (XLSX 10 kb)
Additional file 7:
Details regarding identified Arabidopsis thaliana and apple GASA genes. Gene names and protein sequences were listed. (XLSX 14 kb)
Additional file 8:
Sequences of primers used to amplify MdGASA genes and their reference genes. (DOCX 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Fan, S., Zhang, D., Zhang, L. et al. Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics 18, 827 (2017). https://doi.org/10.1186/s12864-017-4213-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-017-4213-5