Abstract
Background
The vascular bundle in the panicle neck is a crucial trait in rice (Oryza sativa) production that differs between the indica and japonica subspecies. However, the effect of indica/japonica genetic background on the vascular bundles remains unknown.
Results
A series of recombinant inbred lines (RILs) derived from a cross between japonica and indica were planted in three areas. High-throughput sequencing was conducted to determine the indica pedigree percentage and for quantitative trait locus (QTL) analysis. The indica pedigree affected the number of large vascular bundles (LVBs), but not the number of small vascular bundles (SVBs). QTL analysis identified a locus (qLVB9) that was pleiotropic for both LVBs and SVBs in all three areas, and qLVB9 appeared synonymous with DENSE AND ERECT PANICLE 1 (DEP1). Using CRISPR/Cas9 gene editing and gene overexpression technology, we confirmed that the truncated dep1 allele increased the number of LVBs, and resulted in LVBs more closely associated to the indica pedigree. RNA sequencing showed that the truncated dep1 allele downregulated the AP2-like gene family. The double mutant for the DEP1 and AP2-like genes (OsAP2–39) showed decreased endogenous abscisic acid (ABA) level and insensitivity to exogenous ABA treatment, confirming that both DEP1 and OsAP2–39 are involved in the ABA response mechanism.
Conclusions
The present study showed the qLVB9/DEP1 affects LVBs, and involved in ABA signaling via regulating the AP2-like gene family. These results offer new insights into the function of qLVB9/DEP1 in rice.
Similar content being viewed by others
Background
Rice is one of mankind’s major food staples. Due to the continuing growth of the global population and a decrease of availability of arable land, increasing grain yield is an important goal of scientists and rice breeders. In rice breeding, the selection of source, sink, and translocation capacity play critical roles in the improvement of rice yield potential (Donald, 1968, Lafitte and Travis, 1984, Ashraf et al., 1994). The vascular bundle, particularly that in the panicle neck, is the transport system that links the source to the sink and strongly affects the transport efficiency of photosynthetic products, mineral nutrients, and water (Peterson et al., 1982). Vascular bundles in plants play a significant role in the transportation of water, nutrients, and other substances required for biological processes (Lucas et al., 2013). There is a significant positive correlation between grain yield and the number of vascular bundles in rice (Ashraf et al., 1994), wheat (Evans et al., 1970), and oats (Peterson et al., 1982). The capacity of the vascular bundle system to efficiently transport various assimilates has been shown to be a limiting factor for improvements in rice production (Peterson et al., 1982).
The two subspecies of cultivated rice, namely Oryza sativa ssp. indica and O. sativa ssp. japonica, exhibit distinct differences in morphology and ecology. The japonica subspecies is mostly planted in higher latitudes, whereas indica is mainly distributed in lower latitudes (Garris et al., 2005). The vascular bundles are closely related to panicle traits, and there are significant differences of vascular bundles between rice subspecies (Fukuyama et al., 1999). The indica varieties have more vascular bundles in the neck panicles than that in japonica; moreover, the number of vascular bundles is significantly positively correlated with the number of rachis branches (Ling et al., 1982). The V/R (the ratio of the number of large vascular bundles to the number of primary branches) is an important parameter distinguishing indica from japonica, as the V/R is nearly one in japonica and approximately two in indica (Chen et al., 2007).
Quantitative trait locus (QTL) mapping has been utilized to elucidate the genetic mechanisms underlying various important agronomic traits. Over the past serveral decades, several QTLs associated with the responses of vascular bundles in the rice panicle neck have been identified (Sasahara et al., 2010, Zhang et al., 2002, Cui et al., 2003, Bai et al., 2012). Notably, Accumulation of photosystem one 1 (APO1) (Terao et al., 2010) and Narrow leaf 1 (NAL1) (Qi et al., 2008, Fujita et al., 2013) have been reported to participate in vascular bundle improvement.
Hybridization between indica and japonica rice for favorable trait selection has led to the development of high-yielding japonica rice in northern China. The introgression of the indica pedigree into the japonica genetic background significantly increased rice production (Sun et al., 2012). However, the effects of the indica and japonica genetic backgrounds on vascular bundle traits remain unclear. In the present study, the vascular bundle traits of a series of recombinant inbred lines (RILs) derived from a cross between indica and japonica that had been planted in three different areas were investigated. Combined with high-throughput resequencing, the indica pedigree percentage was determined for each RIL, and QTL analysis was conducted to detect the loci that were associated with vascular bundle responses.
Results
Characterization of the vascular bundles of the parent line and RILs from three different areas
To better understand the effect of the indica/japonica genetic background on vascular bundles in rice, we planted a series of 155 RILs derived from a cross between the indica variety “R99” and japonica variety “SN265” in three typical rice cultivation areas, namely Shenyang (SY) (N42°), Jiangsu (JS) (N32°), and Shenzhen (SZ) (N22°). The locations of the cultivation areas are shown in Fig. 1a. Then, we investigated the LVB, SVB, and V/R of the parent lines and RILs in each area. In the parent lines, a significant difference in the LVBs and V/R was observed between SN265 and R99 in all three areas, whereas a significant difference in SVBs between the parent lines was only detected in SZ (Fig. 1b-d). The RILs showed significant differences in LVBs among the three areas. The RILs in JS and SZ showed similar SVBs and V/R distributions, both significantly higher than that in SY (Fig. 1e-g). Analysis of variance (ANOVA) revealed differences among years, areas, and lines. In general, the variation among lines had the largest effect on LVBs, SVBs, and V/R, and significant differences were also observed among the areas, although no significant differences in vascular bundle traits were detected between 2016 and 2017 (Additional file 1: Table S1).
The vascular bundle characteristics of the parent lines and RILs in the three areas. a The locations of the three cultivated areas. b–d The vascular bundle characteristics of SN265 and R99 in the three areas. e–g The vascular bundle characteristics of the RILs in the three areas. * represents significance at the 5% level. h The distribution of the indica pedigree percentage of RILs. i The correlation efficiency of indica pedigree percentage to vascular bundles. The dotted lines indicate significance at the 5% level
The relationship between vascular bundles and yield components
To demonstrate the relationship between vascular bundles and yield components, we conducted a yield component analysis immediately after all of the lines in all three of the areas had reached maturity. The results showed that LVB, SVB, and V/R significantly affected grain number (Table 1). Moreover, LVB also affected the setting rate, whereas SVB was significantly negatively correlated with panicle number. Both the LVB and V/R showed a significant positive correlation with yield (Table 1). V/R regulated grain number in both the primary and secondary branches. Interestingly, LVB and SVB only affected grain number on the secondary branches but not on the primary branches, while LVB also only affected the grain-setting rate on the secondary branches (Table 1).
The effects of the indica/japonica genetic background on vascular bundles
To elucidate the effects of the indica/japonica genetic background on vascular bundles, we conducted high-throughput sequencing of the RILs. The genomes of 155 RIL lines were sequenced to an average approximate depth of 6.25-fold on an Illumina HiSeq 2500 instrument, and the depth of the parent lines reached 30-fold. The high-throughput sequencing detected a total of 1,794,441 SNPs between SN265 and R99. Based on the study of divergence of 517 rice landraces in Huang et al. (Huang et al., 2010), totally 100,529 subspecies-specific SNPs that were the same in all of the japonica samples, but were different in indica were selected. We matched the 100,529 subspecies-specific SNPs to the 1,794,441 SNPs between SN265 and R99, and a total of 61,920 SNPs were merged. The 61,920 SNPs were then used for indica pedigree percentage analysis. In the present study, we defined the indica pedigree percentage as the ratio of the number of indica type SNPs to all 61,920 of the subspecies-specific SNPs, and the results showed that the indica pedigree percentage of the RILs followed a normal distribution (Fig. 1h). We further investigated the relationship between indica pedigree percentage and vascular bundles, the results showed that the indica pedigree percentage was positively correlated to V/R and the number of LVBs in all three of the areas, whereas the effect of the indica pedigree percentage on SVB differed among the three areas, and the correlation was not significant (Fig. 1i). Thus, the indica pedigree percentage mainly affects LVBs.
QTL analysis of the vascular bundles among the three areas
We used the 1,794,441 SNPs between SN265 and R99 for QTL analysis. We namedd the SNPs that co-segregated with one another as “bins,” and a total of 3333 bins were used to construct the molecular linkage map using Highmap software. The phenotypic data of the LVBs, SVBs, and V/R of the three areas were used for the QTL analysis. A total of 17 QTLs for all of the traits were mapped independently on rice chromosomes 1, 2, 3, 6, 7, 8, 9, and 10 (Fig. 2 and Additional file 2: Table S4). Notably, one QTL cluster, qLVB9, was highlighted. The qLVB9 cluster was pleiotropic for both LVBs and SVBs and was detected in all three of the areas. To further identify the exact gene responsible for qLVB9, the number of LVBs in SZ was selected and used in fine-mapping. We mapped the candidate gene to a 43-kb interval in block 19,948 (Fig. 3a). There were seven annotated genes in this bin. One of these genes, DENSE AND ERECT PANICLE 1 (DEP1), has been previously shown to be a regulator for erect panicle architecture, grain number and nitrogen utilization. We subsequently conducted sequence analysis, and found that SN265 had a replacement of a 637-bp stretch in the exon 5 by a 12-bp sequence (Fig. 3a). To verify the effect of DEP1 on vascular bundles, the RILs were further divided into two groups, the DEP1 group (R99 type) and the dep1 group (SN265 type). We found that the number of LVBs and SVBs in the dep1 group was significantly higher than that in the DEP1 group in all three areas. The V/R of the dep1 group was also higher than that of the DEP1 group, but only that in SZ was significant (Fig. 3b and Additional file 3: Table S2). In addition, the correlation between the indica pedigree percentage and the vascular bundle traits in the dep1 group was markedly higher than that in the DEP1 group (Additional file 3: Table S2). Taken together, these findings indicate that DEP1 may be a major pleiotropic locus for both LVBs and SVBs, and furthermore, that the vascular bundles appear to be closely related to the indica pedigree percentage under the dep1 genetic background than the DEP1 genetic background.
The fine mapping of qLVB9 and the function change of qLVB9. a The candidate gene qLVB9 was identified as Os09g0441900/DEP1. b The different functions of the normal DEP1 allele and truncated dep1 allele among three areas, and the correlation efficiency between vascular bundles traits and indica pedigree of these two alleles in three areas. * represents significance at the 5% level, the dotted lines indicate significance at the 5% level
Verification by CRISPR/Cas9 gene editing technology and gene overexpression
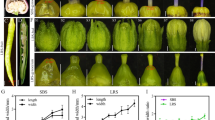
We used CRISPR/Cas9 technology to confirm that DEP1 is a candidate gene for qLVB9 in the japonica cultivar Sasanishiki. The sgRNA was designed in the middle of exon 5, similar to SN265 (Fig. 4a). At least 30 independent regenerated transgenic lines were obtained. We sequenced the T2 mutants, and found five homozygous mutants (Fig. 4b). The deletion in exon 5 of four mutant lines was predicted to result in a frame shift. The sequence substitution in one mutant line was predicted to lead to an amino acid change. The number of LVBs in the four frame shift mutations was significantly increased compared to Sasanishiki, whereas the substitution line showed a similar number of LVBs to that in Sasanishiki (Fig. 4b). The details concerning agronomic traits of WT and CRISPR/Cas9 gene edited plants ware given in Additional file 4: Table S5. We further constructed a DEP1 overexpression plant to verify the function of DEP1 in vascular bundle regulation. The full-length cDNA (SN265) with a 35S promoter was introduced into SN265 (Additional file 5: Figure S1). Among the transgenic lines, TL35 and TL44 exhibited a significant increase in DEP1 expression compared to the wild-type. The number of LVBs in these two lines was also significantly increased (Additional file 5: Figure S1). Thus, we concluded that DEP1 regulates vascular bundle traits.
The CRISPR/Cas9-induced DEP1 mutant lines and LVB investigation. a Schematic map of the genomic region of DEP1 and the sgRNA target site. Arrows show the position of the PCR primers used for mutation detection. b The sequence of the mutant lines and the number of LVBs of the mutant lines. c The WT plant and CRISPR/Cas9 induced DEP1 mutant line. d The difference in vascular bundles between WT plants and the CRISPR/Cas9 induced DEP1 mutant line
RNA-Seq identifies DEP1-regulated genes
To establish the regulatory function of DEP1, RNA-Seq analysis was performed using total RNA extracted from wild-type (Sasanishiki) and CRISPR/Cas9-gene edited plants CRISPR171–21(dep1-crispr) at the heading stage, as CRISPR171–21 had the highest number of LVBs among the mutant lines. High-throughput sequencing using the Illumina HiSeq 2500 platform generated approximately 48.345 Gb of clean data. Furthermore, a total of 76 differentially expressed genes (DEGs) were detected between the wild-type and CRISPR171–21 plants, including 50 that were downregulated and 26 that were upregulated in CRISPR171–21. The Gene Ontology (GO) analysis and hierarchical clustering results of the DEGs are presented in Additional file 6: Table S6 and Additional file 7: Figure S2. Four AP2 (APETALA-2-Like transcription factor gene) genes were enriched in DEGs. We conducted a gene expression analysis for the AP2 genes, and the results showed that Os04g0610400 exhibited the largest difference between CRISPR171–21 and the wild-type among the AP2 genes. Os04g0610400/OsAP2–39 has previously been shown to control abscisic acid (ABA) levels in rice (Yaish et al., 2010). In addition, we subsequently conducted expression profiling of various organs; both DEP1 and OsAP2–39 exhibited the highest expression levels in the rachis meristems (Additional file 8: Figure S3). Thus, we selected OsAP2–39 for the subsequent experiments.
DEP1 involved ABA signaling via OsAP2–39
To confirm the relationship between DEP1 and OsAP2–39, we generated a double mutant for DEP1 and OsAP2–39 using CRISPR/Cas9 technology under the genetic background of CRISPR171–21 (carrying dep1-crispr allele). We obtained three independent mutant lines (CRISPR5060–2, CRISPR5060–3, and CRISPR5060–5), the sequences for which are shown in Fig. 5b. We compared the endogenous ABA levels of the WT, dep1-crspr mutant and dep1-crspr /osap2–39 double mutant. The endogenous ABA level of WT was significant higher than those of dep1-crspr mutant and dep1-crspr /osap2–39 double mutant (Fig. 5d). We then compared the ABA response of DEP1 and OsAP2–39. The seeds of three genotypes, namely the wild-type (DEP1/OsAP2–39), dep1 single mutant CRISPR171–21 (dep1-crspr /OsAP2–39), and double mutant CRISPR5060–2 (dep1-crspr /osap2–39), were sown on sterile filter paper in Petri dishes supplemented with different concentrations of ABA. A significant delay in germination was observed for all of the seeds compared with the control. However, the germination of DEP1/OsAP2–39 was more sensitive to exogenous ABA treatment than dep1-crspr /OsAP2–39 and dep1-crspr /osap2–39. The double mutant dep1-crspr/osap2–39 demonstrated the best germination in the ABA treatment (Additional file 9:Figure S4). In combination, these findings indicate that the truncated dep1 participates in ABA signaling by downregulating AP2-like genes. We then investigated whether ABA was involved in the formation of LVBs. We applied exogenous ABA (10 μM) into irrigation water at the panicle initiation stage (about 35 days before heading) (Yuan et al., 2009). Then we recorded the number of LVBs 3 weeks after ABA application. The results showed that the plants with exogenous ABA applied slightly decreased the number of LVBs compared with the control plants. And the exogenous ABA decreased the expression level of OsAP2–39 compared with control plants (Additional file 10: Figure S5).
The endogenous ABA level and vascular bundles of WT and CRISPR/Cas9-induced DEP1 mutant lines. a Schematic map of the genomic region of OsPA2–39 and the sgRNA target site and the sequences of the mutant lines. The arrows show the position of the PCR primers used for mutation detection. b The sequences of the mutant lines. c The WT plant and CRISPR/Cas9 induced DEP1 mutant line. d The endogenous ABA levels of WT and mutant lines. e The difference in vascular bundles between WT plants and the CRISPR/Cas9 induced DEP1 mutant line
Discussion
Indica and japonica are two subspecies of Asian cultivated rice that can be differentiated in terms of vascular bundle architecture. Indica varieties tend to have more LVBs and a higher V/R ratio. The V/R of indica ranges from 1.6 to 2.0, whereas that of japonica is around 1.0. The increase in V/R ratio in japonica may be a mechanism for improving yield (Fukuyama et al., 1999). Our study confirmed that LVB numbersand V/R in indica-type lines are significantly higher than in japonica-type lines, whereas no distinct differences in SVBs were observed between the two subspecies. In japonica-type cultivars, each of the LVBs is directly connected to a primary branch. In indica-type cultivars, some LVBs are directly connected to the secondary rachis branches because there is more than one LVB in one primary branch (Fukushima and Akita, 1997, Terao et al., 2010). Our study showed that LVBs largely affect the grain number and setting rate of the secondary branches, but not that of the primary branches.
In this study, we detected a pleiotropic QTL (qLVB9) for both LVBs and SVBs in all three areas, and subsequent positional cloning and genetic complementation revealed that qLVB9 is synonymous with DEP1; a gene that has been previously shown to regulate rice panicle architecture and grain number (Huang et al., 2009, Wang et al., 2009). DEP1 has a modular arrangement with a conventional plant-specific Gγ subunit protein domain at its N-terminus, followed by a cysteine-rich domain at the C-terminus (Huang et al., 2009, Sun et al., 2014). G protein signaling participates in various growth and developmental processes in plants and animals (Temple and Jones, 2007, Xu et al., 2016). Thus, variations in G proteins may severely affect different important agronomic traits. However, RNA-Seq analysis has only identified a few DEGs (76 genes) in response to changes in DEP1. Nevertheless, we found AP2-like genes that were downregulated in CRISPR171–21. The overexpression of OsAP2–39 leads to a reduction in yield by decreasing grain number per panicle (Yaish et al., 2010). Thus, the truncated dep1 may increase grain number per panicle by downregulating the expression of OsAP2–39. Moreover, the overexpression of OsAP2–39 caused an increase in the expression of a key ABA biosynthetic gene, OsNCED-1, which subsequently led to an increase in endogenous ABA levels. ABA signaling influences vascular and bundle sheath cells in Arabidopsis (Galvezvaldivieso et al., 2009). Recent studies have demonstrated that OsAP2–39 is also involved in drought tolerance (Wan et al., 2011), and qPE9–1 (also synonymous with DEP1) and RGB1 play distinct roles in ABA responses and drought adaptation (Zhang et al., 2015). Thus, both DEP1 and OsAP2–39 have been shown to be involved in ABA signaling and drought responses. In this study, the ABA treatment for DEP1/OsAP2–39, dep1-crispr/OsAP2–39, and dep1-crispr/osap2–39 plants also demonstrated that both DEP1 and OsAP2–39 are involved in the ABA response mechanism, and these two genes may work in the same pathway.
Previous studies have shown that the dep1 allele results in an increase in grain number and a decrease in setting rate by regulating the Gn1a (Huang et al., 2009, Ashikari et al., 2005). Our study has revealed that the truncated dep1 can significantly increase the number of both LVBs and SVBs. LVBs were significantly positively correlated with grain number and significantly negatively correlated with setting rate (Table 1). These results may explain the phenotype associated with an increase in grain number and a decrease in setting rate in dep1 plants. We determined that plants with a truncated dep1 are significantly more sensitive to indica pedigree percentage than the wild-type DEP1 plants. In dep1 plants, the number of LVBs significantly increased with the increase in indica pedigree percentage, whereas no significant change was observed in the DEP1 plants. Moreover, the indica pedigree percentage was negatively correlated with SVBs in DEP1 plants, but significantly positively correlated with SVBs in dep1 plants. The new function of DEP1 may present a strategy for breeding under the indica genetic background.
In addition to qLVB9/DEP1, our study also detected a QTL (qLVB6) for LVB within the 28.45–28.83 Mb interval on chromosome 6 in JS and SZ. In this region, Os06g0685700, which is similar to the auxin response factor gene, is considered to be the candidate gene of qLVB6. Transgenic plants of Os06g0685700 exhibit pleiotropic defects in growth and development (Huang et al., 2016), and a genome-wide association study also predicted that Os06g0685700 is a LVB-regulating gene (Zhai et al., 2018). Sequence analysis detected several SNPs and InDels at exon regions between SN265 and R99 (Additional file 11: Figure S6), but the function of Os06g0685700 in the vascular bundles requires further investigation. The previous studies demonstrated that APO1 (Terao et al., 2010) and NAL1(Qi et al., 2008, Fujita et al., 2013) are participate in vascular bundle improvement. Our study detected the QTLs corresponding to LVBs at Chr. 6 in JS and SZ, which was consistent with the APO1. However, the NAL1 locus was absent in the QTL analysis of LVB. Then we compare the sequence of NAL1 between SN265 and R99. The results showed that the sequence of NAL1 in SN265 was identical to Nipponbare, and the sequence of NAL1 in R99 was identical to 93–11. We believe that the NAL1 might mainly effect the number and the distribution pattern of LVBs in leaves, or the effect of NAL1 may be weaker under some particular genetic background. Nevertheless, more experiments concerning relationship among NAL1, APO1, DEP1, and OsAP2–39 would be interest.
Conclusions
Vascular bundle is an important trait in rice production, and a highly significant difference was found between the two subspecies, indica and japonica. However, the effect of an indica/japonica genetic background ono the vascular bundles remained unknown. The present study showed that qLVB9/DEP1 has a large effect on the number of LVBs, and the vascular bundles of plants harboring the truncated qLVB9/DEP1 allele were more sensitive to the alteration of ecological conditions and genetic background compared to plants that carried the normal qLVB9/DEP1 allele. Moreover, qLVB9/DEP1 was involved in ABA signaling via regulating the AP2-like gene family. These results offer new insights into the function of qLVB9/DEP1 in rice.
Methods
Plant materials
A total of 155 RILs derived from a cross between ‘Shennong265’ (O. sativa L. ssp. japonica) and ‘R99’ (O. sativa L. ssp. indica) were used in this study. This RIL population was developed from a single-seed descendant that had been inbred for over 10 generations. Field experiments were conducted in three typical rice cultivation areas: the Rice Research Institute of Shenyang Agricultural University (SY) (N41°, E123°), the sub-base of the China National Hybrid Rice R&D Center in Jiangsu Province (JS N32°, E120°), and the Agricultural Genomics Institute at Shenzhen (SZ) (N22°, E114°) for two growing seasons in 2016–2017. Cultivation methods and field management were described in our previous report (Li et al., 2018b) (Additional file 12: Table S3). The paddies were harvested 45 days after heading for each line in each of the three areas.
Evaluation of vascular bundles, yield components and ABA level
We evaluated the vascular bundles after the flowering stage. The main stems with spikes were sampled from eight plants per RIL. The method of vascular bundle evaluation was conducted as described in Liu et al. (2016). At the mature stage (35 days after flowering), the aboveground portions of eight plants per RIL were harvested from each plot. The yield components measurement was conducted as described in Li et al. 2018a (Li et al., 2018b). The evaluation of endogenous level and exogenous ABA response was conducted as described in Yaish et al. (2010).
Genome resequencing and QTL mapping
The genomic DNA of the RILs together with their parents (SN265 and R99) was isolated using the cetyltrimethyl ammonium bromide (CTAB) method (Murray and Thompson, 1980). Sequencing libraries were constructed from these samples and sequenced on the Illumina HiSequation 2500 system according to the manufacturer’s instructions. The detail are described in our previous study (Li et al., 2018a).
Vector construction and plant transformation
We conducted the CRISPR/Cas9 gene editing vector construction as described by (Li et al., 2017). The 23-bp targeting sequences (including PAM) were selected within the target genes, and their targeting specificity was confirmed using a BLAST search against the rice genome (Hsu et al., 2013) Rice transformation was performed as described elsewhere (Nishimura et al., 2006). Genomic DNA was extracted from these transformants, and primer pairs flanking the designed target site were used for PCR amplification. The PCR products (300–500 bp) were sequenced directly and identified using the Degenerate Sequence Decoding method (Ma et al., 2015). The cDNA was cloned into the pBWA(V) HS vector between the 35S promoter and terminator, generating a 35S::DEP1 construct (Additional file 5: Figure S1).
RNA sequencing (RNA-Seq) analysis
Total RNA (10 mg) was extracted from young panicles of the CRISPR171–21 and wild-type plants at the heading stage, and three CRISPR171–21 or wild-type biological samples were pooled together. We conducted the RNA-Seq library preparation and DEGs analysis according to the method described in (Li et al., 2015).
References
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Asuka N, Angeles Enrique R, Qian Q, Hidemi K, Makoto M (2005) Cytokinin oxidase regulates rice grain production. Science 309(5735):741–745
Ashraf Mohammad, Akbar Muhammad, Salim Muhammad, Slafer G. A 1994. Genetic improvement in physiological traits of rice yield. Genetic Improvement of Field Crops.
Bai X, Wu B, Xing Y (2012) Yield-related QTLs and their applications in Rice genetic improvement. J Integr Plant Biol 54(5):300–311
Chen S, Zhengjin X, Wenfu C, Hai X, Hongguang L, Chunjie Z, Yun W, Jiayu W (2007) The relation between the characters of vascular bundle and panicle in the filial generation from Indica and japonica Rice. Acta Agr Boreali-Sinica 5:004
Cui K, Peng S, Xing Y, Yu S, Xu C, Zhang Q (2003) Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor Appl Genet 106(4):649–658
Donald CM (1968) The breeding of crop ideotypes. Euphytica 17(3):385–403
Evans LT, Dunstone RL, Rawson HM, Williams RF (1970) The phloem of the wheat stem in relation to requirements for assimilate by the ear. Aus J Biol Sci 23(4):743–752
Fujita D, Rudi TK, Grubanzo TA, Veronica SM, Yohei K, Kazuhiro S, Nikolaos T, Bueno GR, Takeshi N, Seiji Y (2013) NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. P Natl Acad Sci USA 110(51):20431–20436
Fukushima A, Shigemi A (1997) Varietal differences of the course and differentiation time of large vascular bundles in the rachis of Rice. Jap J Crop Sci 66(1):24–28
Fukuyama T, Sasahara H, Fukuta Y (1999) Variation of vascular bundle system corresponds to indica, tropical-and temperate-japonica differentiation of Asian Rice (Oryza sativa L.). Breeding Sci 49(1):15–19
Galvezvaldivieso G, Fryer Michael J, Tracy L, Katie S, William T, Nicholas S, Asami Tadao DWJ, Jones Alan M, Baker Neil R (2009) The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21(7):2143
Garris Amanda J, Tai Thomas H, Jason C, Steve K, Susan MCOUCH (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169(3):1631–1638
Hsu Patrick D, Scott David A, Weinstein Joshua A, Ann RF, Silvana K, Vineeta A, Li Y, Fine Eli J, Xuebing W, Ophir S (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31(9):827–832
Huang J, Li Z, Dazhong Z (2016) Deregulation of theOsmiR160 target GeneOsARF18Causes growth and developmental defects with an alteration of auxin signaling in Rice. Sci Rep 6(29938):29938
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41(4):494–497
Huang X, Xinghua W, Tao S, Qiang Z, Qi F, Yan Z, Canyang L, Chuanrang Z, Tingting L, Zhiwu Z (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42(11):961–967
Lafitte HR, Travis RL (1984) Photosynthesis assimilate partitioning in closely related lines of Rice exhibiting different sink:source relationships. Crop Sci 24(3)
Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H (2015) Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of Opaque2 in maize. Plant Cell 27(3):532
Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J (2017) A natural allele of a transcription factor in Rice confers broad-Spectrum Blast resistance. Cell 170(1):114–126
Li X, Wu L, Wang J, Sun J, Xia X, Geng X, Wang X, Xu Z, Xu Q (2018a) Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biol 16(1):102
Li X, Wu L, Geng X, Xiuhong X, Wang X, Xu Z, Xu Q (2018b) Deciphering the environmental impacts on Rice quality for different Rice cultivated areas. Rice 11(1):7
Ling QH, Cai JZ, Su ZF (1982) The relationship between the number of macro-vascular bundles in culms and the panicle characters in rice plant and its application. J Jiangsu Agricultural College 3(3):7–16
Liu T, Wenjing B, Jia Z, Yue C, Zhiqiang Y, Yuanzheng W, Cheng F, Hai X, Liang T, Wenfu C (2016) Characterization of the relationship between vascular bundles features and indica -allelic frequency using a seventh filial generations of indica × japonica rice crosses. Euphytica 209(3):1–10
Lucas William J, Andrew G, Raffael L, Kaori F, Ram YS, Yk H, He XQ, Hiroo F, Julie K, Brady Siobhan M (2013) The plant vascular system: evolution, development and functions. J Integr Plant Biol 55(4):294–388
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4325
Nishimura A, Ikuko A, Makoto M (2006) A protocol for agrobacterium-mediated transformation in rice. Nat Protoc 1(6):2796
Peterson DM, Housley TL, Luk TM (1982) Oat stem vascular size in relation to kernel number and weight. II. Field environment 1. Crop Sci 22(2):274–278
Qi J, Qingyun B, Li S, Qian C, Jiaqiang S, Wenxing L, Yihua Z, Chengcai C, Li X, Fugang R (2008) Mutation of the Rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol 147(4):1947
Sasahara H, Yoshimich F, Toshinori F (2010) Mapping of QTLs for vascular bundle system and spike morphology in Rice, Oryza satva L. Breeding Sci 49(2):75–81
Sun H, Qian Q, Kun W, Jijing L, Shuansuo W, Chengwei Z, Yanfei M, Qian L, Xianzhong H, Qingbo Y (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet 46(6):652–656
Sun J, Liu D, Wang J-Y, Ma D-R, Tang L, Gao H, Xu Z-J, Chen W-F (2012) The contribution of intersubspecific hybridization to the breeding of super-high-yielding japonica rice in Northeast China. Theor Appl Genet 125(6):1149–1157
Temple BR, Jones AM (2007) The plant heterotrimeric G-protein complex. Annu Rev Plant Biol 58:249–266
Terao T, Nagata K, Morino K, Hirose T (2010) A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor Appl Genet 120(5):875–893
Wan L, Zhang J, Zhang H, Zhang Z, Quan R, Zhou S, Huang R (2011) Transcriptional activation of OsDERF1 inOsERF3andOsAP2-39Negatively modulates ethylene synthesis and drought tolerance in Rice. PLoS One 6(9):e25216
Wang J, Nakazaki T, Chen S, Chen W, Saito H, Tsukiyama T, Okumoto Y, Xu Z, Tanisaka T (2009) Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.). Theor Appl Genet 119(1):85–91
Xu Q, Zhao M, Wu K, Fu X, Liu Q (2016) Emerging insights into heterotrimeric G protein signaling in plants. J Genet Genomics 43:495–502
Yaish MW, Elkereamy A, Zhu T, Beatty PH, Good AG, Bi YM, Rothstein SJ (2010) The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in Rice. PLoS Genet 6(9):e1001098
Yuan Q, Saito H, Okumoto Y, Inoue H, Nishida H, Tsukiyama T, Teraishi M, Tanisaka T (2009) Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor Appl Genet 119(4):675–684
Zhai L, Tianqing Z, Xinyu W, Yun W, Kai C, Shu W, Jianlong X, Zhikang L (2018) QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice 11(1):13
Zhang D-P, Zhou Y, Jian-Feng Y, Xue-Jiao Y, Sheng L, Wei-Feng X, František B, Yi-Ping W, Yi-Ji X, Guo-hua L (2015) Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J Exp Bot 66(20):6371–6384
Zhang Z-H, Li P, Ling-Xia W, Chang-Jun T, Zhong-Li H, Ying-Guo Z, Li-Huang Z (2002) Identification of quantitative trait loci (QTLs) for the characters of vascular bundles in peduncle related to indica-japonica differentiation in rice (Oryza sativa L.). Euphytica 128(2):279–284
Acknowledgments
The National Natural Science Foundation of China (31430062) supported this study.
Funding
The National Natural Science Foundation of China (31430062) supported this study.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Author information
Authors and Affiliations
Contributions
ZX and QX designed the research plan; CF and XG performed most of the experiments; and QX wrote the paper. All of the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. ANOVA analysis of year, area, and lines for LVBs, SVBs, and V/R (PPTX 65 kb)
Additional file 2:
Table S4. The QTLs information of three areas (PPTX 78 kb)
Additional file 3:
Table S2. The effect of the DEP1/dep1 allele on LVBs, SVBs, and V/R (PPTX 58 kb)
Additional file 4:
Table S5. The agronomic traits of WT and gene edited plants (PPTX 80 kb)
Additional file 5:
Figure S1. The overexpression of DEP1. (a) Overexpression construct for rice transformation. (b) The expression levels of TL35, TL44, and wild-type (WT). (c) The number of LVBs of TL35, TL44, and WT. (d) The endogenous ABA level of WT and dep1 overexpression lines. (e) The WT plant and the dep1 overexpression lines. (f) The difference in vascular bundles in WT plant and dep1 overexpression lines. (PPTX 53 kb)
Additional file 6:
Table S6. The RNA-seq information (PPTX 146 kb)
Additional file 7:
Figure S2. Expression patterns of DEGs. (a) Hierarchical clustering of DEGs. (b) GO analysis of the DEGs between CRISPR171–21 and wild-type plants. (c) The expression level of AP2-like genes in CRISPR171–21 and wild-type plants. (PPTX 3444 kb)
Additional file 8:
Figure S3. DEP1 and OsAP2–39 expression levels in various organs of Sasanishiki. (PPTX 561 kb)
Additional file 9:
Figure S4. Germination investigation under exogenous ABA treatment. (PPTX 52 kb)
Additional file 10:
Figure S5. The number of LVBs between control and ABA treatment plants, and the expression level of OsAP2–39 between control and ABA treatment plants. (PPTX 945 kb)
Additional file 11:
Figure S6. Os06g0685700 sequence differences between SN265 and R99 based on the sequence of SN265. (PPTX 46 kb)
Additional file 12:
Table S3. The fertility of the soil in three areas (PPTX 57 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fei, C., Geng, X., Xu, Z. et al. Multiple areas investigation reveals the genes related to vascular bundles in rice. Rice 12, 17 (2019). https://doi.org/10.1186/s12284-019-0278-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-019-0278-x