Abstract
The cereal plant barley (Hordeum vulgare) has high nutritional content and special characteristics that allow it to withstand adverse weather conditions. The peduncle’s developmental properties under dry (drought and heat) conditions make it an ideal attribute for studying its function in tolerance and adaption processes. Thus, we intend to investigate the natural phenotypic and genetic variation of the peduncle length, as well as the genetic reasons underpinning its growth in dry (drought and heat) environments. Under natural drought and heat, the examined characteristic showed a large range of natural variation, showing its involvement in adaptation to such circumstances. A genome-wide association study (GWAS) was carried out to evaluate the relationship between 127,022 single nucleotide polymorphisms (SNPs) and peduncle length at maturity in 184 barley accessions from throughout the world. The genomic research suggest the possibility of 86 marker-trait associations (p-value: > FDR) in various genomic areas, five of which are physically situated on chromosome 3H. The presence of 23 candidate genes was discovered after predicting candidate genes underlying significant relationships. The RNA-sequencing expression profile of candidate genes revealed that the gene HORVU.MOREX.r2.3HG0216310.1, which was identified as a Polynucleotidyl transferase ribonuclease H-like superfamily protein, had the greatest level of expression, indicating its participation in peduncle development under these conditions. Quantifying the current natural variation in peduncles under dry environmental circumstances and identifying the genomic areas linked with the features will serve as the foundation for future genetic research to better understand its function in stress tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Barley’s (Hordeum vulgare) global production is paramount as it stands as one of the top major crops, serving as a staple food and an essential source of carbohydrates for communities worldwide (Geng et al. 2022). The high demand for cereal plants implies an urgent need to improve both the quality and quantity of crops in order to meet the food security goals for the global population. Nevertheless, the current trends of climate change in arid areas add more necessity to that urgency. Some of the main challenges that need to be confronted to fulfill such demand are overcoming drought and heat stress (Deihimfard et al. 2023). The production of some crop yields such as barley and wheat are foreseen to decrease by 4–7% for each 1 °C increase in temperature above the optimal temperature (Jacott and Boden 2020). Barley is known for its ability to tolerate harsh, arid conditions, especially when compared to its close relative, wheat. Given the predicted scenarios of global warming, research has identified barley as an ideal candidate for studying and potentially enhancing crop production’s adaptation to climate change due to its drought tolerance (Dawson et al. 2015). Such ambition can be met through extensive research that dissects determining barley’s genotypic characteristics, and determining the genetically rich regions and genes responsible for combating drought (Sallam et al. 2019).
Different developmental traits are assumed to contribute to drought tolerance in barley, among which are plant height, root length, and peduncle length (Sallam et al. 2019). The peduncle length is a developmental trait that presents the distance between the last internode connected to the spike. Its length is proportional to plant height; the longer the peduncle, the higher the plant is. Thus, it is believed that the genetic makeup of the two traits contains some similarities (Liu et al. 2023). Both traits are desirable in cereal plants as they contribute to increased crop yield through improving photosynthetic efficiency, and lodging resistance. The peduncle is important in carbon sequence-ration in plants prior to and during grain filling where thereafter carbon is transferred to grains (Kong et al. 2010). Although all internodes could serve as carbohydrate reserves, the peduncle has the highest potential to contribute to the overall photosynthetic productivity in comparison to lower internodes as is exposed to sunlight (Torralbo et al. 2019; Gebbing 2003). Moreover, in a carbon dioxide-stressed environment, the peduncle was found to contribute to the leaf’s photosynthetic productivity (Torralbo et al. 2019).
Understanding the genetic structure of these traits could pave the way for developing molecular tools that are able to enhance crop quality and quantity (e.g. Farooq et al. 2018). The genome-wide association study (GWAS) approach facilitated dissecting the genotypes that influence flowering time (Maurer et al. 2015), plant height (Alqudah et al. 2016), grain weight (Maurer et al. 2016), adaptation to fertilizers and environmental changes (Sharma et al. 2018; Herzig et al. 2018). Many of the associated genes and regions in these investigations showed pleiotropic effects that are linked to more than one trait. Moreover, GWAS has been implemented to improve the efficiency of breeding programs (Genievskaya et al. 2018). There is a limited number of investigations that provide insight into the relationship between genetic markers and peduncle characteristics in barley. Zahn et al. (2021) used GWAS and quantitative trait loci (QTL) to analyze the genetic markers influencing peduncle diameter and tissue thickness. In an experimental setting, two regions in chromosome 5 were found to be associated with peduncle length in barely plants exposed to drought stress, and in some cases, the same QTL region influenced more than one trait such as peduncle length and plant height. Interestingly, it was noted that the same trait can be promoted by different QTLs depending on environmental conditions (Jabbari et al. 2018). Furthermore, manipulating molecular pathways and gene expression in loci responsible for internode length was facilitated by Patil et al. (2019). Here, a disruption in one of the genes responsible for promoting a stronger peduncle is applied through blocking the binding site of microRNA (mi172) in gene APETALA2. Mutated samples had lower mitotic division rates, shorter stem cells, and thus weaker peduncles. Such investigations facilitate the importance of employing genetic research in breeding programs and crop enhancement.
In this study, we aimed to understand the natural variation in the peduncle under harsh conditions and to detect the genetic markers associated with it in response to such conditions. In addition, significant single nucleotide polymorphisms (SNPs) and potential candidate genes were investigated. These results will maximize our genetic knowledge regulating peduncle development and further contribute to the genetic improvement of stress tolerance in barley.
Materials and methods
Data acquisition and Pre-processing
The EcoSeed Spring Barley Collection at IPK-Gatersleben (Germany) was screened under field conditions for peduncle length and plant height. More details on the EcoSeed panel are provided in Alqudah et al. (2021). A Field trial was conducted at the Leibniz Institute of Plant Genetics and Crop Plant Research (Gatersleben, Germany) during the 2018 growing season. This season has been described as the driest growing season for spring barley in middle Germany in decades (Zscheischler and Fischer 2020). Each accession was sown in three independent plots, which comprised 1.2 m2 (1 m long by 1.2 m wide), and contained six rows. Peduncle length and plant height were obtained from more than 10 biological replicates were recorded at maturity. Data distribution was plotted for each trait through a normal probability curve and histogram (supplementary material Fig. 1).
Genotypic information from 184 accessions yielded a total of 127,022 SNPs (Alqudah et al. 2021). This dataset was assembled by combining 4,809 SNPs from the Illumina 9K marker chip and 122,213 SNPs from the Genotyping-By-Sequencing technique (GBS). All used SNPs were subjected to a quality check where Minor Allelic Frequency (MAF) ≥ 5%. Details about the population structure and other molecular characterization of the population were published by Alqudah et al., (2021)
Data analysis
Data in this study were analyzed using R Studio version 2022. Correlation among phenotypic traits and Analysis of Variance (ANOVA) were tested at p < 0.05. The restricted maximum likelihood (REML) algorithm was applied for Best Linear Unbiased Estimators (BLUEs) calculations using the lme4 package (Bates et al., 2015) as described by Alqudah, et al. (2020) to estimate the mean value of each accession over the replicates. The graphics were generated by the R version (RStudio-Team 2015) using the R package ggplot2 (Wickham 2016).
Genome-wide association study approach and candidate genes identification
FarmCPU was utilized in this analysis to ensure the best-fitted model recognizing the association between SNPs and the selected phenotype (Liu et al. 2016). We applied genome-wide analysis among BLUE and SNPs to calculate the associated SNPs with variation in peduncle length and plant height in our test individuals, this statistical approach had been commonly used in barley as described by Alqudah, et al. (2020). Manhattan plots were created by placing markers according to their physical position on the genome on the x-axis in correspondence to their -log10 (p-value) on the y-axis. While quantile–quantile (QQ) plots were constructed showing the expected values on the x-axis and observed values on the y-axis.
SNPs passed the false discovery rate (FDR) threshold of the –log10 (p-value) of SNP ≥ 4.93 were considered as significant associated quantitative trait nucleotides (QTNs) and used for further analysis.
Strong associated SNPs were further used to identify the high-confidence (HC) putative candidate genes underlying the natural variation of the traits within the linkage disequilibrium (LD) interval. The physical position of QTNs and candidate genes were defined using the barley database BARLEX of Morex version 2 (Colmsee et al. 2015).
Barlex database was used for molecular and cellular characterization and gene annotations of the candidate genes (https://apex.ipk-gatersleben.de/apex/f?p=284:10::::::).
Expression analysis for the candidate genes has been done through the RNA‐Seq expression data from the Morex barley cultivar that includes the expression of genes at different stem development stages as TPM (Transcripts Per Kilobase Million).
Results
Phenotypic data
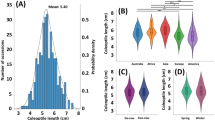
The peduncle length showed a bimodal distribution with two close-by peaks with a mean of 8.61 and wide data distribution (standard deviation = 6.79) indicating wide variability among tested individuals (Fig. 1). Likely, plant height showed broad variability among samples (mean = 21.4, standard deviation = 9.57) and a mono-peak normal distribution (Fig. 1).
Several significantly positive relations were found among phenotypic data with variant strengths of correlation, a strong positive correlation was observed in the correlation matrix between peduncle length and plant height at maturity which was 0.69. None of the tested related traits exhibited a negative correlation with one another. However, some were not significantly associated with each other (Fig. 2).
SNP-based GWAS and candidate gene discovery
The high density of SNPs was concentrated in chromosomes 3, 6, and 7 in locations between 170 to 430 Mb (Fig. 3). The total number of SNPs that are found to be significant (p < 0.05) is 145 distributed in chromosomes 1, 3, 5, 6, and 7 with 1, 131, 5, 5, and 3 SNPs, respectively. We found a total of 9438 genetic markers in chromosome 3. Based on GWAS analysis, five genomic regions with highly significant SNPs were detected to be associated with the traits of interest (Table S1). On the first region, the highest SNP (m_46750) was detected to be significantly associated with peduncle length and mapped on chromosome 3H with p = 2.05E-07 at position 631,337,837 bp which explained positive phenotypic variation by 2.57 (Table S1). In the same sense, the second region showed a significant SNP marker (m_31498) that was found to be associated with peduncle length with p = 6.75E-07 at position 265,648,599 bp (Table S1). Notably, the third genomic region showed the highest number of SNPs where most of the highly significant SNP markers with p-value ≥ 5 (Table S1).
QQ plots of p-values deviated toward the expected axis (Fig. 4). These results suggest the existence of an association between phenotypic and genotypic data in both traits.
Candidate genes
82 genes from the Barlex database Morex V2 that matched our SNPs on chromosome 3H with a high confidence level were identified for peduncle length and 2 for plant height (Table 1). Out of these, we highlighted 22 that contain markers (Table 1). The gene (HORVU.MOREX.r2.3HG0215060.1 encoding Protein translocase subunit SecA) was the only gene that contained three markers, genes (HORVU.MOREX.r2.3HG0215880.1 encoding Kinesin-like protein), (HORVU.MOREX.r2.3HG0212540.1 encoding DNA gyrase subunit B) and (HORVU.MOREX.r2.3HG0212600.1 encoding Ubiquitin-associated domain-containing protein 2), each had two markers while rest of the 18 candidate genes had 1 marker each. We examined the expression profile of each candidate gene and focused on the highly expressed NOD (Developing tillers, 3rd internode—42 DAP) and INF1 (Young developing inflorescences—5 mm) traits for further discussion in their association with the peduncle length and plant height in barley. RNA sequencing of these two traits revealed an expression range between 0.21 to 150.27 FPKM (Fragments Per Kilobase of transcript per Million mapped reads) for NOD and 1.76 to 105.70 FPKM for INF1. Altogether, our potential candidate genes can be used in breeding programs to develop cultivars with the desired stature, thus contributing to improved grain quality and yield under drought and heat-prone environmental conditions.
Discussion
In this study, we used genome-wide association to screen for the association between genetic markers and the peduncle length and plant height in spring barley. A total of 184 accessions were covered with 127,022 genetic markers through the Farm-CPU model. SNPs were well distributed across the genome with condensation in some regions in chromosomes 3H, 6H, and 7H suggesting an important genetic variation in these regions. After generating the Manhattan plot, it was evident that some significant SNPs on chromosome 3H may be associated with the peduncle length and plant height (Fig. 5). When investigating peduncle length, 25 SNPs were detected in this region. Out of these, three had decreased peduncle length while the rest had positive effects. Also, there was only one SNP for plant height that was found in region three (m_30085), the same SNP is shared with peduncle length resulting in decreases of both traits (Supplementary material Table S2). The statistical model used in this study (FarmCPU) suggested a significant correlation between the SNPs identified in 22 candidate genes on chromosome 3H and peduncle length. It should be noted that approximately 62.20% of candidate genes associated with peduncle length are located within region 3 (222,162,361–250,294,215 bp). This supports the previous statement on the existing correlation. Recently, Zahn et al. (2021) reported that important QTLs affecting peduncle length in barley have been located on chromosome 2H, which may influence the transport of nutrients and water to the developing grain, affecting yield and quality. As well, Mora et al. (2016) reported that important QTLs affecting peduncle length in barley have been located on chromosome 2H, which may influence the transport of nutrients and water to the developing grain, affecting yield and quality. For plant height, Niu et al. (2022) reported significant QTLs on chromosome 2H related to plant height, including those that may be linked to the sdw1 dwarfing gene. These genes are of interest for developing shorter, lodging-resistant cultivarss that are easier to harvest and can produce stable yields across different environments (Thabet et al. 2020a, b) (Table 1).
When scanning the expression profile of candidate genes, we targeted two traits that are correlated with the peduncle length, the developing tillers after 42 days (NOD; usually happens 40 days after planting) and young developing inflorescence after 5 mm (INF1). These traits can be used in barley as an adaptation mechanism to harsh conditions by manipulating its developmental cycle to speed up the flowering stage. In this study, the highest expressed gene in NOD and INF1 is polynucleotide transferase ribonuclease H-like superfamily protein with 150.27 and 105.70 FPKM, respectively. This protein family is known for its diverse roles in intracellular functions such as ribonuclease activity, nucleic acid binding, altering RNA, and mechanic responses such as reacting to biotic stimulus, and wound repairing. Polynucleotide transferase ribonucleases (PNTR) are enzymes involved in RNA metabolism, including the processing, editing, and degradation of RNA molecules (Walley et al. 2010a, b). These enzymes can influence various aspects of plant growth and development (Nishanth 2024). PNTR enzymes could affect the expression of genes that regulate peduncle length and plant height (Suzuki et al. 2015a, b).
For instance, genes involved in hormone biosynthesis, signaling pathways (like gibberellins, auxins, and brassinosteroids), and structural components of the cell could be impacted, indirectly influencing these growth traits. These enzymes might play a role in how plants respond to stress (e.g., drought, nutrient deficiency) that can, in turn, affect peduncle growth and overall plant height (Liang et al. 2009a, b). Efficient RNA turnover and processing can be crucial for the rapid adaptation of plants to changing environmental conditions, potentially affecting growth and development (Nishanth 2024). The activity of PNTR enzymes might affect the timing of developmental transitions and the rate of growth, which are critical for determining the final height of the plant and the length of the peduncle (Altmann et al. 2020a, b). Taken these findings together, PNTR is highly expressed under abiotic stress such as drought which is the case in our study.
Another highly expressed gene in NOD and INF1 stages is dentin sialophosphoprotein-like protein with (86.94) and (62.05) FPKM, respectively. The function of this gene is not explored yet in cereal plants, however, in mammals, this gene is essential for biomineralization and teeth hardening development (Yamakoshi 2009; Shang et al. 2023). This aligns with the idea that it is used by barley to resist harsh environmental conditions that place strains on its structural development. Interestingly, DNA gyrase subunit B was under-expressed at NOD (0.21 FPKM) demonstrating the decreased DNA activity under stress conditions and highly expressed at INF1 (88.02 FPKM) where higher genetic activity was needed to support this stage of development. Manipulating molecular features is one of the mechanisms plants usually use to tolerate drought (Razi and Muneer 2023). We found the lowest expression rates at NOD (0.23 FPKM) and INF1 (15.54 FPKM) in Cyclin indicating downregulation when water is scarce. In Arabidopsis thaliana, observed levels of expression of Cyclin were negatively correlated with dehydration levels (Zhou et al. 2013). Another gene that we observed to be under-expressed is the Phosphate transporter (PTs) (19.46 and 11.10 FPKM and NOD and INF1, respectively). It was demonstrated in literature before that, plants’ ability to absorb phosphorus drops over time under prolonged drought, thus, expression of phosphate transporter decreases (Karthikeyan et al. 2002; Bi et al. 2023). Similarly, water scarcity impacts the intracellular ionic profile by interrupting metal transportation activities mirroring the lower expression of gene metal transporter (4.07 and 1.76 FPKM for NOD and INF1, respectively) (Gill et al. 2021). Moreover, PTs might play a role in determining overall plant vigor and stature by ensuring that phosphate is efficiently distributed to growing tissues, including the stem (Cao et al. 2021b, a). PT activity might indirectly affect plant height by modulating the availability of phosphorus for biosynthetic pathways related to growth hormones (Vysotskaya et al. 2020a, b). Understanding the role of PTs in barley development is crucial for breeding programs aimed at improving nutrient use efficiency (Niu et al. 2013a, b). Altogether, cultivars with enhanced phosphate uptake and transport capabilities could maintain optimal growth and development, including desirable peduncle length and plant height, even under stress conditions.
Remarkably, there were two members of the Zinc finger CCCH domain protein on chromosome 3H but none of which was expressed in the developing tillers or inflorescence stages. This gene family is known for its significant role in drought tolerance in plants, hence, it controls several molecular pathways to reduce plant’s sensitivity to dehydration (Ai et al. 2022). Perhaps, the lack of expression of these genes is due to the high drought tolerance in barley and not reaching the adequate stress level to trigger zinc finger CCCH expression, although there were other molecular signs of drought resistance. This emphasizes the need to further dissect chromosome 3 to locate more genotypic traits associated with the peduncle length and plant height in stress conditions in particular.
Conclusion
Our GWAS analysis revealed a strong association between peduncle length and SNPs in chromosome 3H. Further observations into candidate genes showed a strong alliance with RNA sequences of specific traits. Selected traits were linked to the flowering stage in barley directly and indirectly to stress tolerance. Our study is in favor of supposing a strong association between peduncle length and specific genotypic traits. We suggest further investigations to elaborate on how the expression profile of these genotypes influences peduncle length through different developmental stages.
References
Ai Q, Pan W, Zeng Y, Li Y, Cui L (2022) CCCH Zinc finger genes in Barley: genome-wide identification, evolution, expression and haplotype analysis. BMC Plany Biology 22:117. https://doi.org/10.1186/s12870-022-03500-4
Alqudah AM, Koppolu R, Wolde GM, Graner A, Schnurbusch T (2016) The genetic architecture of barley plant stature. Front Genet 7:117. https://doi.org/10.3389/fgene.2016.00117
Alqudah AM, Sallam A, Baenziger PS, Börner A (2020) GWAS: fast-forwarding gene identification and characterization in temperate Cereals: lessons from Barley—a review. J Adv Res 22:119–135
Alqudah AM, Sharma R, Börner A (2021) Insight into the genetic contribution of maximum yield potential, spikelet development and abortion in barley. Plants People Planet 3:721–736. https://doi.org/10.1002/ppp3.10203
Altmann M, Altmann S, Rodriguez PA, Weller B, Elorduy Vergara L, Palme J, Marín-de la Rosa N, Sauer M, Wenig M, Villaécija-Aguilar JA, Sales J, Lin CW, Pandiarajan R, Young V, Strobel A, Gross L, Carbonnel S, Kugler KG, Garcia-Molina A, Bassel GW, Falter C, Mayer KFX, Gutjahr C, Vlot AC, Grill E, Falter-Braun P (2020a) Extensive signal integration by the phytohormone protein network. Nature 583:271–276
Altmann M, Altmann S, Rodriguez PA, Weller B, Vergara LE, Palme J, Falter-Braun P (2020b) Extensive signal integration by the phytohormone protein network. Nature 583(7815):271–276. https://doi.org/10.1038/s41586-020-2460-0
Bates D, Maechler M, Bolker B, Walker S, Christensen RHB, Singmann H, Bolker MB (2015) Package ‘lme4.’ Convergence 12(1):2
Bi W, Zhang D, Weng B, Dong Z, Wang F, Wang W, Yan D (2023) Research progress on the effects of droughts and floods on phosphorus in soil-plant ecosystems based on knowledge graph. HydroResearch 6:29–35
Cao Y, Liu J, Li Y, Zhang J, Li S, An Y, Tianming H, Yang P (2021a) Functional analysis of the phosphate transporter gene MtPT6 from medicago truncatula. Front Plant Sci. https://doi.org/10.3389/fpls.2020.620377
Cao Y, Liu J, Li Y, Yang P (2021b) Functional analysis of the phosphate transporter gene MtPT6 from Medicago truncatula. Front Plant Sci 11:620377
Colmsee C, Beier S, Himmelbach A, Schmutzer T, Stein N, Scholz U, Mascher M (2015) BARLEX–the barley draft genome explorer. Mol Plant 8(6):964–966
Dawson TP, Russell J, Powell W, Steffenson B, Thomas WTB, Waugh R (2015) Barley: a translational model for adaptation to climate change. N Phytol 206:913–931
Deihimfard R, Rahimi-Moghaddam S, Eyni-Nargeseh H, Collins B (2023) An optimal combination of sowing date and cultivar could mitigate the impact of simultaneous heat and drought on rainfed wheat in arid regions. Eur J Agron 147:126848. https://doi.org/10.1016/j.eja.2023.126848
Farooq MU, Cheema AA, Ishaaq I, Zhu J (2018) Correlation and genetic component studies for peduncle length affecting grain yield in wheat. Int J Adv Appl Sci 5(10):67–75. https://doi.org/10.21833/ijaas.2018.10.010
Gebbing T (2003) The enclosed and exposed part of the peduncle of wheat (Triticum aestivum)–spatial separation of fructan storage. New Phytol 159(1):245–252
Geng L, Li M, Zhang G, Ye L (2022) Barley: a potential cereal for producing healthy and functional foods. Food Qual Safe. https://doi.org/10.1093/fqsafe/fyac012
Genievskaya Y, Almerekova S, Sariev B, Chudinov V, Tokhetova L, Sereda G et al (2018) Marker-trait associations in two-rowed spring barley accessions from Kazakhstan and the USA. PLoS ONE 13(10):e0205421
Gill RA, Ahmar S, Ali B, Saleem MH, Khan MU, Zhou W, Liu S (2021) The Role of membrane transporters in plant growth and development, and abiotic stress tolerance. Int J Mol Sci 22(23):12792. https://doi.org/10.3390/ijms222312792
Herzig P, Maurer A, Draba V, Sharma R, Draicchio F, Bull H, Milne L, Thomas WTB, Flavell AJ, Pillen K (2018) Contrasting genetic regulation of plant development in wild barley grown in two European environments revealed by nested association mapping. J Exp Bot 69(7):1517–1531. https://doi.org/10.1093/jxb/ery002
Jabbari M, Fakheri BA, Aghnoum R, Mahdi Nezhad N, Ataei R (2018) GWAS analysis in spring barley (Hordeum vulgare L.) for morphological traits exposed to drought. PLoS ONE 13(9):e0204952
Jacott CN, Boden SA (2020) Feeling the heat: developmental and molecular responses of wheat and barley to high ambient temperatures. J Exp Bot 71(19):5740–5751. https://doi.org/10.1093/jxb/eraa326
Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130(1):221–233. https://doi.org/10.1104/pp.020007
Kong L, Wang F, Feng B, Li S, Si J, Zhang B (2010) The structural and photosynthetic characteristics of the exposed peduncle of wheat (Triticum aestivum L.): an important photosynthate source for grain-filling. BMC Plant Biol 10:1–10
Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X, Li C (2009a) The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 19:307–316
Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Li C (2009b) The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 19(3):307–316
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767
Liu Y, Shen K, Yin C, Xiaowan X, Xuchang Y, Ye B, Sun Z, Dong J, Bi A, Zhao X, Daxing X, He Z, Zhang X, Hao C, Jianhui W, Wang Z, He W, Liu D, Zhang L, Shen L, Hao Y, Fei L, Guo Z (2023) Genetic basis of geographical differentiation and breeding selection for wheat plant architecture traits. Genom Biol. https://doi.org/10.1186/s13059-023-02932-x
Maurer A, Draba V, Jiang Y, Schnaithmann F, Sharma R, Schumann E, Kilian B, Reif JC, Pillen K (2015) Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genom 16:290. https://doi.org/10.1186/s12864-015-1459-7
Maurer A, Draba V, Pillen K (2016) Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. J Exp Bot 67(8):2507–2518. https://doi.org/10.1093/jxb/erw070
Mora F, Quitral YA, Matus I, Russell J, Waugh R, Del Pozo A (2016) SNP-based QTL mapping of 15 complex traits in barley under rain-fed and well-watered conditions by a mixed modeling approach. Front Plant Sci 7:187078
Nishanth MJ (2024a) Transcriptome meta-analysis-based identification of hub transcription factors and RNA-binding proteins potentially orchestrating gene regulatory cascades and crosstalk in response to abiotic stresses in Arabidopsis thaliana. J Appl Genet 65(2):255–269. https://doi.org/10.1007/s13353-024-00837-4
Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS (2013a) Responses of root architecture development to low phosphorus availability: a review. Ann Bot 112:391–408
Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS (2013b) Responses of root architecture development to low phosphorus availability: a review. Ann Bot 112(2):391–408
Niu Y, Chen T, Zheng Z, Zhao C, Liu C, Jia J, Zhou M (2022) A new major QTL for flag leaf thickness in barley (Hordeum vulgare L.). BMC Plant Biol. https://doi.org/10.1186/s12870-022-03694-7
Patil V, McDermott HI, Mcallister T, Cummins M, Clara Silva J, Mollison E, Meikle R, Morris JA, Hedley PE, Waugh R, Dockter C, Hansson M, McKim S (2019) APETALA2 control of barley internode elongation. Development 146(11):170373. https://doi.org/10.1242/dev.170373
Razi K, Muneer S (2023) Grafting enhances drought tolerance by regulating and mobilizing proteome, transcriptome and molecular physiology in okra genotypes. Front Plant Sci 14:1178935. https://doi.org/10.3389/fpls.2023.1178935
Sallam A, Alqudah A, Dawood M, Baenziger PS, Börner A (2019) Drought stress tolerance in wheat and barley: advances in physiology, breeding and genetics research. Int J Mol Sci 20(13):3137. https://doi.org/10.3390/ijms20133137
Shang B, Wang L, Yan X, Li Y, Li C, Wu C, Zhang X (2023) Intrinsically disordered proteins SAID1/2 condensate on SERRATE for dual inhibition of miRNA biogenesis in Arabidopsis. Proc Natl Acad Sci 120(14):e2216006120
Sharma R, Draicchio F, Bull H, Herzig P, Maurer A, Pillen K, Thomas WTB, Flavell AJ (2018) Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. J Exp Bot 69:3811–3822
Suzuki Y, Arae T, Green PJ, Yamaguchi J, Chiba Y (2015a) AtCCR4a and AtCCR4b are involved in determining the poly(A) length of granule-bound starch synthase 1 transcript and modulating sucrose and starch metabolism in Arabidopsis thaliana. Plant Cell Physiol 56:863–874
Suzuki Y, Arae T, Green PJ, Yamaguchi J, Chiba Y (2015b) AtCCR4a and AtCCR4b are involved in determining the poly (A) length of granule-bound starch synthase 1 transcript and modulating sucrose and starch metabolism in Arabidopsis thaliana. Plant Cell Physiol 56(5):863–874
Team R (2015) RStudio: integrated development for R. RStudio. Inc., Boston, MA
Thabet SG, Moursi YS, Karam MA, Börner A, Alqudah AM (2020a) Natural variation uncovers candidate genes for barley spikelet number and grain yield under drought. Stress 11:533
Thabet SG, Moursi YS, Karam MA, Börner A, Alqudah AM (2020b) Natural variation uncovers candidate genes for barley spikelet number and grain yield under drought stress. Genes 11(5):533
Torralbo F, Vicente R, Morcuende R, González-Murua C, Aranjuelo I (2019) C and N metabolism in barley leaves and peduncles modulates responsiveness to changing CO2. J Exp Bot 70(2):599–611
Vysotskaya L, Akhiyarova G, Feoktistova A, Akhtyamova Z, Korobova A, Ivanov I, Dodd I, Kuluev B, Kudoyarova G (2020a) Effects of phosphate shortage on root growth and hormone content of barley depend on capacity of the roots to accumulate ABA. Plants 9(12):1722. https://doi.org/10.3390/plants9121722
Vysotskaya L, Akhiyarova G, Feoktistova A, Akhtyamova Z, Korobova A, Ivanov I, Kudoyarova G (2020b) Effects of phosphate shortage on root growth and hormone content of barley depend on capacity of the roots to accumulate ABA. Plants 9(12):1722
Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K (2010a) Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 152:866–875
Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K (2010b) Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 152(2):866–875
Wickham H (2016) Data analysis. Springer International Publishing, New York, pp 189–201
Yamakoshi Y (2009) Dentinogenesis and dentin sialophosphoprotein (DSPP). J Oral Biosci 51(3):134. https://doi.org/10.2330/joralbiosci.51.134
Zahn S, Schmutzer T, Pillen K, Maurer A (2021) Genomic dissection of peduncle morphology in barley through nested association mapping. Plants 10:10. https://doi.org/10.3390/plants10010010
Zhou XF, Jin YH, Yoo CY, Lin XL, Kim WY, Yun DJ, Bressan RA, Hasegawa PM, Jin JB (2013) CYCLIN H;1 regulates drought stress responses and blue light-induced stomatal opening by inhibiting reactive oxygen species accumulation in Arabidopsis. Plant Physiol 162(2):1030–1041. https://doi.org/10.1104/pp.113.215798
Zscheischler J, Fischer EM (2020) The record-breaking compound hot and dry 2018 growing season in Germany. Weather Clim Extremes 29:100270
Acknowledgements
The authors (ZK and AMA) thank Qatar University for providing the required analysis platforms for the project. Open Access funding is provided by the Qatar National Library. AMA and AB would extend their thanks to IPK-Gatersleben for internal funding supporting this study.
The author (MDA) would like to acknowledge Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2024R355), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding
Open Access funding provided by the Qatar National Library. The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
AMA designed the experiment. AMA, SS, CK, and EL did the field work and phenotyping. ZK and AMA analyzed the data. ZK, SGT, MDA, MS, AB, and AMA wrote and edited the manuscript. ZK, SG, MS, and AMA conceived the idea and participated in the interpretation of the results. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
I declare that the authors have no competing interests that might be perceived to influence the results and/or discussion reported in this paper.
Ethical approval
This article does not contain any research involving animals or human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khatir, Z., Thabet, S.G., Alqahtani, M.D. et al. Discovery of new genomic regions and candidate genes implicated in the natural variation of barley peduncle length and plant height. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02050-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02050-y