Abstract
Key message
A core marker set containing markers developed to be informative within a single commercial cotton species can elucidate diversity structure within a multi-species subset of the Gossypium germplasm collection.
Abstract
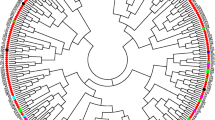
An understanding of the genetic diversity of cotton (Gossypium spp.) as represented in the US National Cotton Germplasm Collection is essential to develop strategies for collecting, conserving, and utilizing these germplasm resources. The US collection is one of the largest world collections and includes not only accessions with improved yield and fiber quality within cultivated species, but also accessions possessing sources of abiotic and biotic stress resistance often found in wild species. We evaluated the genetic diversity of a subset of 272 diploid and 1,984 tetraploid accessions in the collection (designated the Gossypium Diversity Reference Set) using a core set of 105 microsatellite markers. Utility of the core set of markers in differentiating intra-genome variation was much greater in commercial tetraploid genomes (99.7 % polymorphic bands) than in wild diploid genomes (72.7 % polymorphic bands), and may have been influenced by pre-selection of markers for effectiveness in the commercial species. Principal coordinate analyses revealed that the marker set differentiated interspecific variation among tetraploid species, but was only capable of partially differentiating among species and genomes of the wild diploids. Putative species-specific marker bands in G. hirsutum (73) and G. barbadense (81) were identified that could be used for qualitative identification of misclassifications, redundancies, and introgression within commercial tetraploid species. The results of this broad-scale molecular characterization are essential to the management and conservation of the collection and provide insight and guidance in the use of the collection by the cotton research community in their cotton improvement efforts.





Similar content being viewed by others
References
Abdalla AM, Reddy OUK, El-Zik KM, Pepper AE (2001) Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor Appl Genet 102:222–229
Abdurakhmonov IY, Kohel RJ, Yu JZ, Pepper AE, Abdullaev AA, Kushanov FN, Salakhutdinov IB, Buriev ZT, Saha S, Scheffler BE, Jenkins JN, Abdukarimov A (2008) Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics 92:478–487
Alvarez I, Wendel JF (2006) Cryptic interspecific introgression and genetic differentiation within Gossypium aridum (Malvaceae) and its relatives. Evolution 60:505–517
Blenda A, Fang DD, Rami JF, Garsmeur O, Luo F, Lacape JM (2012) A high density consensus genetic map of tetraploid cotton that integrates multiple component maps through molecular marker redundancy check. PLoS One 7:e45739
Campbell BT, Williams VE, Park W (2009) Using molecular markers and field performance data to characterize the Pee Dee cotton germplasm resources. Euphytica 169:285–301
Campbell BT, Saha S, Percy R, Frelichowski J, Jenkins JN, Parka W, Mayee CD, Gotmare V, Dessauw D, Giband M, Du X, Jia Y, Constable G, Dillon S, Abdurakhmonov IY, Abdukarimov A, Rizaeva SM, Abdullaev A, Barroso PAV, Pádua JG, Hoffmann LV, Podolnaya L (2010) Status of the global cotton germplasm resources. Crop Sci 50:1161–1179
Cordeiro GM, Pan YB, Henry RJ (2003) Sugarcane microsatellites for the assessment of genetic diversity in sugarcane germplasm. Plant Sci 165:181–189
Cronn R, Wendel JF (2003) Cryptic trysts, genomic mergers, and plant speciation. New Phytol 161:133–142
Fang DD, Xiao J, Canci PC, Cantrell RG (2010) A new SNP haplotype associated with blue disease resistance gene in cotton (Gossypium hirsutum L.). Theor Appl Genet 120:943–953
Fang DD, Hinze LL, Percy RG, Li P, Deng D, Thyssen G (2013) A microsatellite-based genome-wide analysis of genetic diversity and linkage disequilibrium in Upland cotton (Gossypium hirsutum L.) cultivars from major cotton-growing countries. Euphytica 191:391–401
Frelichowski JE Jr, Palmer MB, Main D, Tomkins JP, Cantrell RG, Stelly DM, Yu J, Kohel RJ, Ulloa M (2006) Cotton genome mapping with new microsatellites from Acala ‘Maxxa’ BAC-ends. Mol Genet Gen 275:479–491
Fryxell PA (1992) A revised taxonomic interpretation of Gossypium L. (Malvaceae). Rheedea 2:108–165
Guo WZ, Cai CP, Wang CB, Han ZG, Song XL, Wang K, Niu XW, Wang C, Lu KY, Shi B, Zhang TZ (2007) A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176:527–541
Guo W, Cai C, Wang C, Zhao L, Wang L, Zhang T (2008) A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genom 9:314
Han ZG, Guo WZ, Song XL, Zhang TZ (2004) Genetic mapping of EST-derived microsatellites from the diploid Gossypium arboreum in allotetraploid cotton. Mol Genet Gen 272:308–327
Han Z, Wang C, Song X, Guo W, Gou J, Li C, Chen X, Zhang T (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Hinze LL, Dever JK, Percy RG (2012) Molecular variation among and within improved cultivars in the U.S. cotton germplasm collection. Crop Sci 52:222–230
Hoffman SM, Yu JZ, Grum DS, Xiao JH, Kohel RJ, Pepper AE (2007) Identification of 700 new microsatellite loci from cotton (Gossypium hirsutum). J Cotton Sci 11:208–241
IBPGR (1980) Descriptors for cotton species. In: International Board for Plant Genetic Resources Working Group, 1979, Rome
Jaccard P (1908) Nouvelles rescherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Jena SN, Srivastava A, Rai KM, Ranjan A, Singh SK, Nisar T, Srivastava M, Bag SK, Mantri S, Asif MH, Yadav HK, Tuli R, Sawant SV (2012) Development and characterization of genomic and expressed SSRs for levant cotton (Gossypium herbaceum L.). Theor Appl Genet 124:565–576
Kantartzi SK, Ulloa M, Sacks E, Stewart JM (2009) Assessing genetic diversity in Gossypium arboreum L. cultivars using genomic and EST-derived microsatellites. Genetica 136:141–147
Kohel R, Yu J, Park Y-H, Lazo G (2001) Molecular mapping and characterization of traits controlling fiber quality in cotton. Euphytica 121:163–172
Kuester AP, Nason JD (2012) Microsatellite loci for Gossypium davidsonii (Malvaceae) and other D-genome, Sonoran Desert endemic cotton species. Am J Bot 99:e91–e93
Lacape JM, Nguyen TB, Thibivilliers S, Bojinov B, Courtois B, Cantrell RG, Burr B, Hau B (2003) A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46:612–626
Lacape JM, Dessauw D, Rajab M, Noyer JL, Hau B (2007) Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol Breed 19:45–58
Liu Q, Brubaker CL, Green AG, Marshall DR, Sharp PJ, Singh SP (2001) Evolution of the FAD2-1 fatty acid desaturase 5′ UTR intron and the molecular systematics of Gossypium (Malvaceae). Am J Bot 88:92–102
Liu D, Guo X, Lin Z, Nie Y, Zhang X (2006) Genetic diversity of Asian cotton (Gossypium arboreum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol 53:1145–1152
Nguyen TB, Giband M, Brottier P, Risterucci AM, Lacape JM (2004) Wide coverage of the tetraploid cotton genome using newly developed microsatellite markers. Theor Appl Genet 109:167–175
Oliveira KM, Pinto LR, Marconi TG, Mollinari M, Ulian EC, Chabregas SM, Falco MC, Burnquist W, Garcia AA, Souza AP (2009) Characterization of new polymorphic functional markers for sugarcane. Genome 52:191–209
Page JT, Gingle AR, Udall JA (2013) PolyCat: a resource for genome categorization of sequencing reads from allopolyploid organisms. G3 Genes Genomes Genet 3:517–525
Park YH, Alabady MS, Ulloa M, Sickler B, Wilkins TA, Yu J, Stelly DM, Kohel RJ, El-Shihy OM, Cantrell RG (2005) Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line cotton population. Mol Genet Gen 274:428–441
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J et al (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Percival AE (1987) The National Collection of Gossypium Germplasm. In: Southern cooperative series bulletin no. 321
Reddy OUK, Pepper AE, Abdurakmonov I, Saha S, Jenkins JN, Brooks T, El-Zik KM (2001) New dinucleotide and trinucleotide microsatellite marker resources for cotton genome research. J Cotton Sci 5:103–113
Reif JC, Melchinger AE, Frisch M (2005) Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci 45:1–7
Rohlf FJ (2000) NTSYSpc: numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software, Setauket
Rungis D, Llewellyn D, Dennis ES, Lyon BR (2005) Simple sequence repeat (SSR) markers reveal low levels of polymorphism between cotton (Gossypium hirsutum L.) cultivars. Aust J Agric Res 56:301–307
Silva DC, Costa Duarte Filho LS, Dos Santos JM, de Souza Barbosa GV, Almeida EC (2012) DNA fingerprinting based on simple sequence repeat (SSR) markers in sugarcane clones from the breeding program RIDESA. Afr J Biotech 11:4722–4728
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Toll JA (1995) Processing of germplasm, associated material and data. In: Guarino L, Ramanatha RV, Reid R (eds) Collecting plant genetic diversity: technical guidelines. CABI, Wallingford, pp 577–595
van Treuren R, de Groot EC, Boukema IW, van de Wiel CCM, van Hintum TJL (2010) Marker-assisted reduction of redundancy in a genebank collection of cultivated lettuce. Plant Genet Resour Charact Util 8:95–105
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu Y-X, Wang J, Yu S (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44:1098–1104
Wang Z, Zhang D, Wang X, Tan X, Guo H, Paterson AH (2013) A whole-genome DNA marker map for cotton based on the D-genome sequence of Gossypium raimondii L. G3 Gene Genome Genet 3:1759–1767
Weir B (1996) Genetic data analysis II: methods for discrete population genetic data. Sinauer Associates Inc, Sunderland
Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78:139–186
Wendel JF, Stewart JM, Rettig JH (1991) Molecular evidence for homoploid reticulate evolution among Australian species of Gossypium. Evolution 45:694–711
Xiao J, Wu K, Fang DD, Stelly DM, Yu J, Cantrell RG (2009) New SSR markers for use in cotton (Gossypium spp.) improvement. J Cotton Sci 13:75–157
Yu Y, Yuan DJ, Liang SG, Li XM, Wang XQ, Lin ZX, Zhang XL (2011) Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genom 12:15
Yu JZ, Fang DD, Kohel RJ, Ulloa M, Hinze LL, Percy RG, Zhang J, Chee P, Scheffler BE, Jones DC (2012a) Development of a core set of SSR markers for the characterization of Gossypium germplasm. Euphytica 187:203–213
Yu JZ, Kohel RJ, Fang DD, Cho J, Van Deynze A, Ulloa M, Hoffman SM, Pepper AE, Stelly DM, Jenkins JN, Saha S, Kumpatla SP, Shah MR, Hugie WV, Percy RG (2012b) A high-density simple sequence repeat and single nucleotide polymorphism genetic map of the tetraploid cotton genome. G3 Gene Genome Genet 2:43–58
Zhang T, Qian N, Zhu X, Chen H, Wang S, Mei H, Zhang Y (2013) Variations and transmission of QTL alleles for yield and fiber qualities in upland cotton cultivars developed in China. PLoS One 8:e57220
Zhao L, Yuanda L, Caiping C, Xiangchao T, Xiangdong C, Wei Z, Hao D, Xiuhua G, Wangzhen G (2012) Toward allotetraploid cotton genome assembly: integration of a high-density molecular genetic linkage map with DNA sequence information. BMC Genom 13:539
Acknowledgments
The authors wish to acknowledge the contributions of S. Simpson of the USDA-ARS Genomics and Bioinformatics Research Unit who managed the whole process of the PCRs from setting up and proofing the plates to monitoring the data output. Without Ms. Simpson’s efforts and diligence this project could not have been completed. The authors also recognize the contributions of members of the USDA-ARS cotton breeding, genetics, and genomics project teams for their valuable technical assistance. This research project was supported by funding from Cotton Incorporated and USDA CRIS Nos. 6202-21000-031-00D (L.H., J.Y., J.F., R.P.), 6435-21000-016-00D (D.F.), 5347-21000-011-00D (M.G.), and 6402-21310-003-00D (B.S.). Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors note that this research is performed and reported in accordance with the ethical standards for scientific conduct in the United States of America.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Alan H. Schulman.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hinze, L.L., Fang, D.D., Gore, M.A. et al. Molecular characterization of the Gossypium Diversity Reference Set of the US National Cotton Germplasm Collection. Theor Appl Genet 128, 313–327 (2015). https://doi.org/10.1007/s00122-014-2431-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2431-7