Abstract
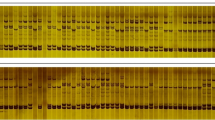
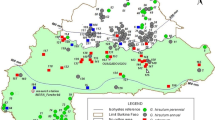
Asian cotton (Gossypium arboreum L.) was once widely cultivated in China. It has also been a valuable source of genetic variation in modern cotton improvement. In this study, the genetic diversity of selected G. arboreum accessions collected from different regions of China was evaluated by microsatellite (simple sequence repeats, SSRs) analysis. Of the 358 microsatellite markers analyzed, 74 primer pairs detected 165 polymorphic DNA fragments among 39 G. arboreum accessions examined. Twelve accessions could be fingerprinted with one or more SSR markers. With the exception of two accessions, DaZiJie and DaZiMian, genetic similarity coefficients among all accessions ranged from 0.58 to 0.87 suggesting high level of genetic variation in the G. arboreum collections. The UPGMA dendrogram constructed from genetic similarity coefficients revealed positive correlation between cluster groupings and geographic distances. In addition, comparison of the microsatellite amplification profiles of the diploid G. arboreum and tetraploid Gossypium hirsutum L. found that size distribution of amplified products in G. arboreum was dispersive and that of G. hirsutum was relatively concentrated. The information on the genetic diversity and SSR fingerprinting from this study is useful for developing mapping populations for constructing diploid cotton genetic linkage map and tagging economically important traits.
Similar content being viewed by others
References
A.M. Abdalla, O.U.K. Reddy, K.M. El-Zik and A.E. Pepper, Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor. Appl. Genet. 102 (2001) 222-229
J.O. Beasley, The origin of American tetraploid Gossypium species. Am. Nat. 74 (1940) 285-286
J.O. Beasley, Meiotic chromosome behavior in species, species hybrids, haploids and induced polyploids of Gossypium. Genetics 27 (1942) 25-54
D.U. Gerstel, Chromosomal translocations in interspecific hybrids of the genus Gossypium. Evolution 7 (1953) 234-244
O.A. Gutiérrez, S. Basu, S. Saha, J.N. Jenkins, D.B. Shoemaker, C.L. Cheatham and J.C. McCarty Jr., Genetic distance among selected cotton genotypes and its relationship with F2 performance. Crop Sci. 42 (2002) 1841-1847
M.J. Iqbal, O.U.K. Reddy, K.M. El-Zik and A.E. Pepper, A genetic bottleneck in the 'evolution under domesticationȁ9 of upland cotton Gossypium hirsutum L. examined using DNA fingerprinting. Theor. Appl. Genet. 103 (2001) 547-554
C.X. Jiang, R.J. Wright, K.M. El-Zik and A.H. Paterson, Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. USA 95 (1998) 4419-4424
S.A. Khan, D. Hussain, E. Askari, J.McD. Stewart, K.A. Malik and Y. Zafar, Molecular phylogeny of Gossypium species by DNA fingerprinting. Theor. Appl. Genet. 101 (2000) 931-938
J.M. Lacape, T.B. Nguyen, S. Thibivilliers, B. Bojinov, B. Courtois, R.G. Cantrell, B. Burr and B. Hau, A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46 (2003) 612-626
S. Liu, S. Saha, D. Stelly, B. Burr and R.G. Cantrell, Chromosomal assignment of microsatellite loci in cotton. J. Hered. 91 (2000) 326-332
H.J. Lu and G.O. Myers, Genetic relationships and discrimination of ten influential Upland cotton varieties using RAPD markers. Theor. Appl. Genet. 105 (2002) 325-331
S.S. Mehetre, A.R. Aher, V.L. Gawande, V.R. Patil and A.S. Mokate, Induced polyploidy in Gossypium: a tool to overcome interspecific incompatibility of cultivated tetraploid and diploid cottons. Curr. Sci. 84 (2003) 1510-1512
M. Mei, N.H. Syed, W. Gao, P.M. Thaxton, C.W. Smith, D.M. Stelly and Z.J. Chen, Genetic mapping and QTL analysis of fiber–related traits in cotton (Gossypium). Theor. Appl. Genet. 108 (2004) 280-291
J.V. Monte, C.L. McIntyre and J.P. Gustafson, Analysis of phylogenetic relationships in the Triticeae tribe using RFLPs. Theor. Appl. Genet. 86 (1993) 649-655
M. Morgante, M. Hanafey and W. Powell, Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30 (2002) 194-200
M. Nei and W.H. Li, Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76 (1979) 5269-5273
A.H. Paterson, C.L. Brubaker and J.F. Wendel, A rapid method for extraction of cotton (Gossypium spp.) genome DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11 (1993) 122-127
F.J. Rohlf, NTSYS-pc: Numerical Taxonomy and Multivariate Analysis SystemVersion 2.1, User Guide. New York: Exeter Software (2000).
T. Seelanan, A. Schnabel and J.F. Wendel, Congruence and consensus in the cotton tribe. Syst. Bot. 22 (1997) 259-290
A. Sharma, R. Sharma and H. Machii, Assessment of genetic diversity in a Morus germplasm collection using fluorescence-based AFLP markers. Theor. Appl. Genet. 101 (2000) 1049-1055
R.R. Sokal and C.D. Michener, A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 28 (1958) 1409-1438
N.H. Syed, H.S. Lee, M. Mei, P. Thaxton, D.M. Stelly and Z.J. Chen, Variability and evolution of microsatellite loci in cotton (Gossypium) diploid and polyploidy genomes. San DiegoCA: Plant & Animal Genome IX Conference. Town & Country Hotel (2001).
V. Tatineni, R.G. Cantrell and D.D. Davis, Genetic diversity in elite cotton germplasm determined by morphological characteristics and RAPDs. Crop Sci. 36 (1996) 186-192
G.V. Vergara and S.S. Bughrara, AFLP analyses of genetic diversity in Bentgrass. Crop Sci. 43 (2003) 2162-2171
X.L. Xiang and D.Z. Shen, Chinese Asian Cotton (Gossypium arboreum). Beijing, China: China Agricultural Press (1989).
Q.H. Xu, X.L. Zhang and Y.C. Nie, Genetic diversity evaluation of cultivars (G. hirsutum L.) from the Changjiang River valley and Yellow River valley by RAPD markers. Acta Genet. Sin. 28 (2001) 683-690
L.F. Zhu, X.L. Zhang and Y.C. Nie, Analysis of genetic diversity in upland cotton (Gossypium hirsutum L.) cultivars from China and foreign countries by RAPDs and SSRs. J. Agric. Biotechnol. 11 (2003) 450-455
Author information
Authors and Affiliations
Corresponding author
Additional information
Diqiu Liu, Xiaoping Guo: These two authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, D., Guo, X., Lin, Z. et al. Genetic Diversity of Asian Cotton (Gossypium arboreum L.) in China Evaluated by Microsatellite Analysis. Genet Resour Crop Evol 53, 1145–1152 (2006). https://doi.org/10.1007/s10722-005-1304-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-005-1304-y