Abstract
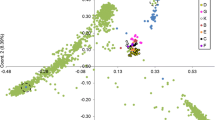
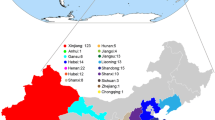
The diploid cotton species Gossypium arboreum possesses many favorable agronomic traits such as drought tolerance and disease resistance, which can be utilized in the development of improved upland cotton cultivars. The USDA National Plant Germplasm System maintains more than 1600 G. arboreum accessions. Little information is available on the genetic diversity of the collection thereby limiting the utilization of this cotton species. The genetic diversity and population structure of the G. arboreum germplasm collection were assessed by genotyping-by-sequencing of 375 accessions. Using genome-wide single nucleotide polymorphism sequence data, two major clusters were inferred with 302 accessions in Cluster 1, 64 accessions in Cluster 2, and nine accessions unassigned due to their nearly equal membership to each cluster. These two clusters were further evaluated independently resulting in the identification of two sub-clusters for the 302 Cluster 1 accessions and three sub-clusters for the 64 Cluster 2 accessions. Low to moderate genetic diversity between clusters and sub-clusters were observed indicating a narrow genetic base. Cluster 2 accessions were more genetically diverse and the majority of the accessions in this cluster were landraces. In contrast, Cluster 1 is composed of varieties or breeding lines more recently added to the collection. The majority of the accessions had kinship values ranging from 0.6 to 0.8. Eight pairs of accessions were identified as potential redundancies due to their high kinship relatedness. The genetic diversity and genotype data from this study are essential to enhance germplasm utilization to identify genetically diverse accessions for the detection of quantitative trait loci associated with important traits that would benefit upland cotton improvement.





Similar content being viewed by others
References
Ahmad S, Mahmood K, Hanif M, Nazeer W, Malik W, Qayyum A, Hanif K, Mahmood A, Islam N (2011) Introgression of cotton leaf curl virus-resistant genes from Asiatic cotton (Gossypium arboreum) into upland cotton (G. hirsutum). Genet Mol Res 10:2404–2414
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379
Erpelding JE, Stetina SR (2013) Genetics of resistance to Rotylenchulus reniformis in Gossypium arboreum accession PI529728. World J Agric Res 1:48–53
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Felsenstein J (1989) PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164–166
Filippi CV, Aguirre N, Rivas JG, Zubrzycki J, Puebla A, Cordes D, Moreno MV, Fusari CM, Alvarez D, Heinz RA, Hopp HE, Paniego NB, Lia VV (2015) Population structure and genetic diversity characterization of a sunflower association mapping population using SSR and SNP markers. BMC Plant Biol 15:52
Gill MS, Bajaj YPS (1987) Hybridization between diploid (Gossypium arboreum) and tetraploid (Gossypium hirsutum) cotton through ovule culture. Euphytica 36:625–630
Girma G, Hyma KE, Asiedu R, Mitchell SE, Gedil M, Spillane C (2014) Next-generation sequencing based genotyping, cytometry and phenotyping for understanding diversity and evolution of guinea yams. Theor Appl Genet 127:1783–1794
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9:e90346
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015:431487
Guo WZ, Zhou BL, Yang LM, Wang W, Zhang TZ (2006) Genetic diversity of landraces in Gossypium arboreum L. race sinense assessed with simple sequence repeat markers. J Integr Plant Biol 48:1008–1017
He J, Zhao X, Laroche A, Lu Z-X, Liu H, Li Z (2014) Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci 5:484. doi:10.3389/fpls.2014.00484
Hedin PA, Jenkins JN, Parrott WL (1992) Evaluation of flavonoids in Gossypium aboreum (L) cottons as potential source of resistance to tobacco budworm. J Chem Ecol 18:105–114
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Iqbal MA, Abbas A, Zafar Y, Rahman MU (2015) Characterization of indigenous Gossypium arboreum L. genotypes for various fiber quality traits. Pak J Bot 47:2347–2354
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Kantartzi SK, Ulloa M, Sacks E, Stewart JM (2009) Assessing genetic diversity in Gossypium arboreum L. cultivars using genomic and EST-derived microsatellites. Genetica 136:141–147
Kebede H, Burow G, Dani RG, Allen RD (2007) A-genome cotton as a source of genetic variability for upland cotton (Gossypium hirsutum). Genet Resour Crop Evol 54:885–895
Kujur A, Bajaj D, Upadhyaya HD, Das S, Ranjan R, Shree T, Saxena MS, Badoni S, Kumar V, Tripathi S, Gowda CLL, Sharma S, Singh S, Tyagi AK, Parida SK (2015) A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea. Sci Rep 5:11166
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li C, Waldbieser G, Bosworth B, Beck BH, Thongda W, Peatman E (2014a) SNP discovery in wild and domesticated populations of blue catfish, Ictalurus furcatus, using genotyping-by-sequencing and subsequent SNP validation. Mol Ecol Resour 14:1261–1270
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, Xiao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2014b) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46:567–572
Lin M, Cai S, Wang S, Liu S, Zhang G, Bai G (2015) Genotyping-by-sequencing (GBS) identified SNP tightly linked to QTL for pre-harvest sprouting resistance. Theor Appl Genet 128:1385–1395
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Lischer HE, Excoffier L (2012) PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299
Liu D, Guo X, Lin Z, Nie Y, Zhang X (2006) Genetic diversity of Asian cotton (Gossypium arboreum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol 53:1145–1152
Logan-Young CJ, Yu JZ, Verma SK, Percy RG, Pepper AE (2015) SNP discovery in complex allotetraploid genomes (Gossypium spp., Malvaceae) using genotyping by sequencing. Appl. Plant Sci 3:1400077. doi:10.3732/apps.1400077
Maqbool A, Abbas W, Rao AQ, Irfan M, Zahur M, Bakhsh A, Riazuddin S, Husnain T (2010) Gossypium arboreum GHSP26 enhances drought tolerance in Gossypium hirsutum. Biotechnol Prog 26:21–25
Marchini J, Howie B (2010) Genotype imputation for genome-wide association studies. Nat Rev Genet 11:499–511
Mehetre SS, Aher AR, Gawande VL, Patil VR, Mokate AS (2003) Induced polyploidy in Gossypium: a tool to overcome interspecific incompatibility of cultivated tetraploid and diploid cottons. Cur Sci 84:1510–1512
Pace J, Gardner C, Romay C, Ganapathysubramanian B, Lübberstedt T (2015) Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genom 16:47
Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Gen 5:92–102
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rahman M, Yasmin T, Tabbasam N, Ullah I, Asif M, Zafar Y (2008) Studying the extent of genetic diversity among Gossypium arboreum L. genotypes/cultivars using DNA fingerprinting. Genet Resour Crop Evol 55:331–339
Robinson AF (2007) Reniform in U.S. cotton: when, where, why, and some remedies. Annu Rev Phytopathol 45:263–288
Romay MC, Millard MJ, Glaubitz JC, Peiffer JA, Swarts KL, Casstevens TM, Elshire RJ, Acharya CB, Mitchell SE, Flint-Garcia SA, McMullen MD, Holland JB, Buckler ES, Gardner CA (2013) Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol 14:R55. doi:10.1186/gb-2013-14-6-r55
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Sacks EJ, Robinson AF (2009) Introgression of resistance to reniform nematode (Rotylenchulus reniformis) into upland cotton (Gossypium hirsutum) from Gossypium arboreum and a G. hirsutum/Gossypium aridum bridging line. Field Crop Res 112:1–6
Samad H, Coll F, Preston MD, Ocholla H, Fairhurst RM, Clark TG (2015) Imputation-based population genetics analysis of Plasmodium falciparum malaria parasites. PLoS Genet 11:e1005131
Stanton MA, Stewart JM, Tugwell NP (1992) Evaluation of Gossypium arboreum L. germplasm for resistance to thrips. Genet Resour Crop Evol 39:89–95
Stanton MA, Stewart JM, Percival AE, Wendel JF (1994) Morphological diversity and relationships in the A-genome cottons, Gossypium arboreum and G. herbaceum. Crop Sci 34:519–527
Thyssen GN, Fang DD, Turley RB, Florane C, Li P, Naoumkina M (2015) Mapping-by-sequencing of Ligon-lintless-1 (Li 1 ) reveals a cluster of neighboring genes with correlated expression in developing fibers of upland cotton (Gossypium hirsutum L.). Theor Appl Genet 128:1703–1712
Ulukan H (2011) Plant genetic resources and breeding: current scenario and future prospects. Int J Agric Biol 13:447–454
Wallace TP, Bowman D, Campbell BT, Chee P, Gutierrez OA, Kohel RJ, McCarty J, Myers G, Percy R, Robinson F, Smith W, Stelly DM, Stewart JM, Thaxton P, Ulloa M, Weaver DB (2009) Status of the USA cotton germplasm collection and crop vulnerability. Genet Resour Crop Evol 56:507–532
Wallace JG, Upadhyaya HD, Vetriventhan M, Buckler ES, Hash CT, Ramu P (2015) The genetic makeup of a global barnyard millet germplasm collection. Plant Gen. doi:10.3835/plantgenome2014.10.0067
Wong MML, Gujaria-Verma N, Ramsay L, Yuan HY, Caron C, Diapari M, Vandenberg A, Bett KE (2015) Classification and characterization of species within the genus Lens using genotyping-by-sequencing (GBS). PLoS One 10:e0122025
Yik CP, Birchfield W (1984) Resistant germplasm in Gossypium species and related plants to Rotylenchulus reniformis. J Nematol 16:146–153
Acknowledgments
This research was funded by the USDA-ARS project 6066-22000-074-00D. This research was supported in part by an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the US Department of Energy (DOE) and the US Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-AC05-06OR23100. All opinions expressed in this paper are the author’s and do not necessarily reflect the policies and views of USDA, ARS, DOE, or ORAU/ORISE. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest in the reported research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2016_9921_MOESM2_ESM.tiff
Supplementary material 2 Fig. 1 Distribution of (a) locus level missing data rate, (b) minor allele frequency, and (c) heterozygosity for the initial 61,822 SNPs, preprocessed 18,571 SNPs, and discriminating 6224 SNPs. (d) Taxa level missing data rate for the 6224 discriminating SNPs (TIFF 97 kb)
10709_2016_9921_MOESM3_ESM.tiff
Supplementary material 3 Fig. 2 Plot of Delta K for (a) all 375 Gossypium arboreum accessions indicating two major clusters, (b) 302 Cluster 1 accessions showing two sub-clusters, and (c) 64 Cluster 2 accessions showing three sub-clusters (TIFF 360 kb)
10709_2016_9921_MOESM4_ESM.tiff
Supplementary material 4 Fig. 3 Population structure at different clustering level for the 6224 discriminating SNPs (TIFF 228 kb)
10709_2016_9921_MOESM5_ESM.tiff
Supplementary material 5 Fig. 4 Distribution of (a) missing data rate, (b) minor allele frequency, and (c) heterozygosity at the locus level for the 375 Gossypium arboreum accessions, 302 Cluster 1, and 64 Cluster 2 accessions (TIFF 3021 kb)
Rights and permissions
About this article
Cite this article
Li, R., Erpelding, J.E. Genetic diversity analysis of Gossypium arboreum germplasm accessions using genotyping-by-sequencing. Genetica 144, 535–545 (2016). https://doi.org/10.1007/s10709-016-9921-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-016-9921-2