Abstract
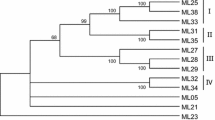
Plant R genes confer resistance to pathogens in a gene-for-gene mode. Seventy-five putative resistance gene analogs (RGAs) containing conserved domains were cloned from Rubus idaeus L. cv. ‘Latham’ using degenerate primers based on RGAs identified in Rosaceae species. The sequences were compared to 195 RGA sequences identified from five Rosaceae family genera. Multiple sequence alignments showed high similarity at multiple nucleotide-binding site (NBS) motifs with homology to Drosophila Toll and mammalian interleukin-1 receptor (TIR) and non-TIR RNBSA-A motifs. The TIR sequences clustered separately from the non-TIR sequences with a bootstrap value of 76%. There were 11 clusters each of TIR and non-TIR type sequences of multiple genera with bootstrap values of more than 50%, including nine with values of more than 75% and seven of more than 90%. Polymorphic sequence characterized amplified region and cleaved amplified polymorphic sequence markers were developed for nine Rubus RGA sequences with eight placed on a red raspberry genetic linkage map. Phylogenetic analysis indicated four of the mapped sequences share sequence similarity to groupTIR I, while three others were spread in non-TIR groups. Of the 75 Rubus RGA sequences analyzed, members were placed in five TIR groups and six non-TIR groups. These group classifications closely matched those in 12 of 13 studies from which these sequences were derived. The analysis of related DNA sequences within plant families elucidates the evolutionary relationship and process involved in pest resistance development in plants. This information will aid in the understanding of R genes and their proliferation within plant genomes.









Similar content being viewed by others
References
Aarts MGM, te Lintel Hekkert B, Holub EB, Beynon JL, Stiekema WJ, Pereira A (1998) Identification of R-gene homologous DNA fragments genetically linked to resistance loci in Arabidopsis thaliana. Mol Plant Microbe Interact 11:252–258
Afunian MR, Goodwin PH, Hunter DM (2006) Search for molecular markers linked to fire blight resistance in pear (Pyrus communis). Acta Hort (ISHS) 704:557–566
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP (1997) Signaling in plant–microbe interactions. Science 276:726–733
Bent AF (1996) Plant disease resistance genes: function meets structure. Plant Cell 8:1757–1771
Baldi P, Patocchi A, Zini E, Toller C, Velaso R, Komjanc M (2004) Cloning and linkage mapping of resistance gene homologues in apple. Theor Appl Genet 109:231–239
Barritt BH, Crandall PC, Bristow PR (1979) Breeding for root rot resistance in red raspberry. J Amer Soc Hort Sci 104:92–94
Calenge F, Van der Linden CG, Van de Weg E, Schouten HJ, Van Arkel G, Denance C, Durel C-E (2005) Resistance gene analogues identified through the NBS-profiling method map close to major genes and QTL for disease resistance in apple. Theor Appl Genet 110:660–668
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant-Microb Interact 11:968–978
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411:826–833
Deng Z, Huang S, Ling P, Chen C, Yu C, Weber CA, Moore GA, Gmitter FG Jr (2000) Cloning and characterization of NBS-LRR class resistance-gene candidate sequences in Citrus. Theor Appl Genet 101:814–822
Donald TM, Pellerone F, Adam-Blondon AF, Bouquet A, Thomas MR, Dry IB (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor Appl Genet 104:610–618
Erwin DC, Ribeiro OK (1996) Phytophthora disease worldwide. APS Press, St. Paul, MN, p 562
Felsenstein J (2006) PHYLIP (Phylogeny Inference Package; http://evolution.genetics.washington.edu/phylip.html)
Flor HH (1971) Current status for the gene-for-gene concept. Ann Rev Phytopathol 9:275–296
Gaspero G, Cipriani G (2002) Resistance gene analogs are candidate markers for disease-resistance genes in grape (Vitis spp.). Theor Appl Genet 106:163–172
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Ann Rev Plant Physiol Plant Mol Biol 48:575–607
Hattendorf A (2005) Molekulargenetische Charakterisierung von Resistenzgenanaloga im Rosengenom als Basis fuer die Resistenzzuechtung. Dept Hort, Hannover, Germany, p 117 University of Hannover
Hattendorf A, Debener T (2007) Molecular characterization of NBS-LRR-RGAs in the rose genome. Physiol Plant 129(4):775–786
Jones JDG (1996) Plant disease resistance genes: structure, function and evolution. Curr Opin Biotechnol 7:155–160
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Karplus K, Barrett C, Hughey R (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics 14(10):846–856
Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using codominant ecotype-specific markers. Plant J 4:403–410
Lalli DA, Decroocq V, Blenda AV, Schurdi-Levraud V, Garay L, Le Gall O, Damsteegt V, Reighard GL, Abbott AG (2005) Identification and mapping of resistance gene analogs (RGAs) in Prunus: a resistance map for Prunus. Theor Appl Genet 111:1504–1513
Lee SY, Lee DH (2003) Putative TIR-NBS type R protein 4 [Malus baccata]: expression of the MbR4, a TIR-NBS type of apple R gene, confers resistance to bacterial spot disease in Arabidopsis. GenBank accessions http://www.ncbi.nlm.nih.gov
Lee SY, Lee DH (2006) TIR-NBS-LRR type R protein 7 [Malus baccata]: overexpression of the Apple MbR7 Encoding a TIR-NBS-LRR Type of R gene Induce Enhanced Resistance of transgenic Arabidopsis Infection with Bacterial Pathogen. GenBank accessions http://www.ncbi.nlm.nih.gov
Lee S-Y, Seo J-S, Rodriquez-Lanetty M, Lee D-H (2003) Comparative analysis of superfamilies of NBS-encoding disease resistance gene analogs in cultivated and wild apple species. Mol Gen Genomics 269:101–108
Leister D, Ballova A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with a potential wide application in plants. Nat Genet 14:421–429
Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P (1998) Rapid reorganization of resistance gene homologues in cereal genome. Proc Natl Acad Sci USA 95:370–375
Liang FS, Kong FN, Zhou C-J, Cao P-X, Ye C-J, Wang B (2005) Cloning and characterization of a non-TIR-NBS-LRR type disease resistance gene analogue from peach. DNA Sequence 16:103–110
Martínez Zamora MG, Castagnaro AP, Díaz Ricci JC (2004) Isolation and diversity analysis of resistance gene analogues (RGAs) from cultivated and wild strawberries. Molec Genet Genomics 272:480–487
McDowell JM, Dhandaydham M, Long TA, Aatrs MG, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10:1861–1874
Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Paran I, Michelmore RW (1993) Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet 85:985–993
Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50:203–213
Pattison JA, Samuelian SK, Weber CA (2007) Inheritance of Phytophthora root rot resistance in red raspberry determined by generation means and molecular linkage analysis. Theor Appl Genet 115:225–236
Radwan O, Bouzidi MF, Nicolas P, Mouzeyar S (2004) Development of PCR markers of the PI5/PI8 locus for resistance to Plasmopara halstedii in sunflower, Helianthus annuus L. from complete CC-NBS-LRR sequences. Theor Appl Genet 109:176–185
Rehm BHA (2001) Bioinformatic tools for DNA/protein sequence analysis, functional assignment of genes and protein classification. Appl Microbiol Biotechnol Online article at http://link.springer.de/link/service/journals/00253/index.htm
Saraste M, Sibbald PR, Wittinghofer A (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Tr Biochem Sci 15:430–434
Shen KA, Meyers BC, Islam-Faridi MN, Chin DB, Stelly DM, Michelmore RW (1998) Resistance-gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol Plant Microbe Interact 11:1815–823
Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley KC (1996) BCM Search Launcher—An integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res 6:454–462
Soriano JM, Vilanova S, Romero C, Llácer G, Badenes ML (2005) Characterization and mapping of NBS-LRR resistance gene analogs in apricot (Prunus armeniaca L.). Theor Appl Genet 110:980–989
Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400:667–671
Tan H, Callahan FE, Zhang X-D, Karaca M, Saha S, Jenkins JN, Creech RG, Ma D-P (2003) identification of resistance gene analogs in cotton (Gossypium hirsutum L.). Euphytica 134:1–7
Timmerman-Vaughan GM, Frew TJ, Weeden NF (2000) Characterization and linkage mapping of R-gene analogous DNA sequences in pea (Pisum sativum L.). Theor Appl Genet 101:241–247
Van Oijen JW, Voorrips RE (2001) JoinMap 3.0®, software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands
Varshochi A (2006) Putative CC-NBS-LRR resistance protein [Malus × domestica]: Sequencing of a Malus domestica BAC associated with resistance to Dysaphis devecta. GenBank accessions. http://www.ncbi.nlm.nih.gov
Wilcox WF, Pritts MP, Kelly MJ (1999) Integrated control of Phytophthora root rot of red raspberry. Plant Dis 83:1149–1154
Xu Q, Wen X, Deng X (2005) Isolation of TIR and nonTIR NBS-LRR resistance gene analogues and identification of molecular markers linked to a powdery mildew resistance locus in chestnut rose (Rosa roxburghii Tratt). Theor Appl Genet 111:819–830
Xu Q, Wen X, Deng X (2007) Phylogenetic and evolutionary analysis of NBS-encoding genes in Rosaceae fruit crops. Mol Phylogenet Evol 44:315–324
Yu YG, Buss GR, Maroof MAS (1996) Isolation of a superfamily of candidate disease resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Zhang LP, Lin GY, Niño-Liu D, Foolad MR (2003) Mapping QTLs conferring early blight (Alternaria solani) resistance in a Lycopersicon esculentum × L. hirsutum cross by selective genotyping. Mol Breeding 12:3–19
Acknowledgments
This research was supported by a grant from the US Department of Agriculture—National Research Initiative (USDA-NRI) NYG-632526. We are grateful to William Boone for skillful technical work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: J. Davis
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table S1
List of all 270 sequences (DOC 423 KB).
S2
Alignment of all 270 amino acid sequences in a format suitable for input into the Phylip phylogenetic inference package (ZIP 23.9 KB)
Supplemental Fig. S3
Detailed tree in color showing all 270 terminal nodes (PDF 46.5 KB)
Supplemental Table S4
Clusters containing multiple genera with a bootstrap of more than 50% and correspondence with other studies (DOC 64 KB)
Rights and permissions
About this article
Cite this article
Samuelian, S.K., Baldo, A.M., Pattison, J.A. et al. Isolation and linkage mapping of NBS-LRR resistance gene analogs in red raspberry (Rubus idaeus L.) and classification among 270 Rosaceae NBS-LRR genes. Tree Genetics & Genomes 4, 881–896 (2008). https://doi.org/10.1007/s11295-008-0160-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-008-0160-2