Abstract
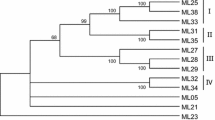
Genomic DNA sequences sharing homology with the NBS-LRR (nucleotide binding site-leucine-rich repeat) resistance genes were isolated and cloned from apricot (Prunus armeniaca L.) using a PCR approach with degenerate primers designed from conserved regions of the NBS domain. Restriction digestion and sequence analyses of the amplified fragments led to the identification of 43 unique amino acid sequences grouped into six families of resistance gene analogs (RGAs). All of the RGAs identified belong to the Toll-Interleukin receptor (TIR) group of the plant disease resistance genes (R-genes). RGA-specific primers based on non-conserved regions of the NBS domain were developed from the consensus sequences of each RGA family. These primers were used to develop amplified fragment length polymorphism (AFLP)-RGA markers by means of an AFLP-modified procedure where one standard primer is substituted by an RGA-specific primer. Using this method, 27 polymorphic markers, six of which shared homology with the TIR class of the NBS-LRR R-genes, were obtained from 17 different primer combinations. Of these 27 markers, 16 mapped in an apricot genetic map previously constructed from the self-pollination of the cultivar Lito. The development of AFLP-RGA markers may prove to be useful for marker-assisted selection and map-based cloning of R-genes in apricot.





Similar content being viewed by others
References
Aarts MGM, Hekkert BL, Holub EB, Beynon JL, Stiekema WJ, Pereira A (1998) Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant-Microbe Interact 11:251–258
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baker B, Zambryski P, Staskewicz B, Dinesh-Kumar SP (1997) Signaling in plant-microbe interactions. Science 276:726–733
Baldi P, Patocchi A, Zini E, Toller C, Velasco R, Komjanc M (2004) Cloning and linkage mapping of resistance gene homologues in apple. Theor Appl Genet 109(1):231–239
Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giradaut J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265:1856–1860
Bliss FA, Arulsekar S, Foolad MR, Becerra V, Gillen AM, Warburton ML, Dandekar AM, Kocsisne GM, Mydin KK (2002) An expanded genetic linkage map of Prunus based on an interspecific cross between almond and peach. Genome 45:520–529
Cordero JC, Skinner DZ (2002) Isolation from alfalfa of resistance gene analogues containing nucleotide binding sites. Theor Appl Genet 104:1283–1289
Deng Z, Huang S, Ling P, Chen C, Yu C, Weber CA, Moore GA, Gmiter FG Jr (2000) Cloning and characterization of NBS-LRR class resistance-gene candidate sequences in citrus. Theor Appl Genet 101:814–822
Di Gaspero G, Cipriani G (2002) Resistance gene analogs are candidate markers for disease-resistance genes in grape (Vitis spp.). Theor Appl Genet 106:163–172
Dilbirligi M, Erayman M, Sandhu D, Sidhu D, Gill KS (2004) Identification of wheat chromosomal regions containing expressed resistance genes. Genetics 166:461–481
Dixon MSK, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84:451–459
Donald TM, Pellerone F, Adam-Blondon AF, Bouquet A, Thomas MR, Dry IB (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor Appl Genet 104:610–618
Doyle JJ, Doyle JL (1987) A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem Bul 19:11–15
Egea-Gilabert C, Dickinson MJ, Bilotti G, Candela ME (2003) Isolation of resistance gene analogs in pepper using modified AFLPs. Biol Plant 47:27–32
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Foulongne M, Pascal T, Pfeiffer F, Kervella J (2003) QTLs for powdery mildew resistance in peach×Prunus davidiana crosses: consistency across generations and environments. Mol Breed 12:33–50
Gentzbittel L, Mouzeyar S, Badaoui S, Mestries E, Vear F, De Labrouhe DT, Nicolas P (1998) Cloning of molecular markers for disease resistance in sunflower, Helianthus annus L.. Theor Appl Genet 96:519–525
Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269:843–846
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Hayes AJ, Saghai Maroof MA (2000) Targeted resistance gene mapping in soybean using modified AFLPs. Theor Appl Genet 100:1279–1283
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Hurtado MA, Romero C, Vilanova S, Abbott AG, Llácer G, Badenes ML (2002) Genetic linkage maps of two apricot cultivars (Prunus armeniaca L.), and mapping of PPV (sharka) resistance. Theor Appl Genet 105:182–191
Initiative TA (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–816
Johal GS, Briggs SP (1992) Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258:985–987
Kanazin V, Frederick Marek L, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kobe B, Deisenhofer J (1994) The leucine-rich repeat: a versatile binding motif. Trends Biol Sci 19:415–421
Kosambi D (1944) The estimation of map distances from recombination value. Ann Eugen 12:172–175
Kuhn DN, Heath M, Wisser RJ, Meerow A, Brown JS, Lopes U, Schnell RJ (2003) Resistance gene homologues in Theobroma cacao as useful genetic markers. Theor Appl Genet 107:191–202
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) mega: molecular evolutionary genetics analysis, version 2.1. Pennsylvania State University, Pittsbergh, Pa.
Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7:1195–1206
Leister D, Ballvora A, Salamini S, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P (1998) Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA 95:370–375
Lescot M, Rombauts S, Zhang J, Aubourg S, Mathé C, Jansson S, Rouzé P, Boerjan W (2004) Annotation of a 95-Kb Populus deltoides genomic sequence reveals a disease resistance gene cluster and novel class I and II transposable elements. Theor Appl Genet 109:10–22
Mago R, Nair S, Mohan M (1999) Resistance gene analogues from rice: cloning, sequencing and mapping. Theor Appl Genet 99:50–57
Martin GB, Brommonschenkel SH, Chunwongse J, Grary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1436
Martínez Zamora MG, Castagnaro AP, Díaz Ricci JC (2004) Isolation and diversity analysis of resistance genes analogues (RGAs) from cultivated and wild strawberries. Mol Genet Genomics 272:480–487
McDowell JM, Dhandaydham M, Long TA, Aarts MGM, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10:1861–1874
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Mindrinos M, Katagiri F, Yu G-L, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78:1089–1099
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M, Kumar S (2002) Molecular evolution and phylogenetics. Oxford University Press, New York
Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50:203–213
Parker JE, Coleman MJ, Szabó V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JDG (1997) The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Interleukin-1 receptors with N and L6. Plant Cell 9:879–894
Quint M, Mihaljevic R, Dussle CM, Xu ML, Melchinger AE, Lübberstedt T (2002) Development of RGA-CAPS markers and genetic mapping of candidate genes for sugarcane mosaic virus resistance in maize. Theor Appl Genet 105:355–363
Quint M, Dussle CM, Melchinger AE, Lübberstedt T (2003) Identification of genetically linked RGAs by BAC screening in maize and implications for gene cloning, mapping and MAS. Theor Appl Genet 106:1171–1177
Rohlf FJ (1993) ntsys-pc numerical taxonomy and multivariate analysis system, version 1.8 Exeter Publications, Setauket
Rozen H, Skaletsky S (2000) primer3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols in the series methods in molecular biology. Humana Press, Totowa, pp 365–386
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salava J, Wang Y, Krska B, Polak J, Kominek P, Miller W, Dowler W, Reighard GL, Abbott AG (2002) Molecular genetic mapping in apricot. Czech J Genet Plant Breed 38:65–68
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Saraste M, Sibbad PR, Wittinghofer A (1990) The P-loop-a common motif in ATP- and GTP- binding proteins. Trends Biochem Sci 15:430–434
Seah S, Sivasithamparam K, Karakousis A, Lagudah ES (1998) Cloning and characterisation of a family of disease resistance gene analogs from wheat and barley. Theor Appl Genet 97:937–945
Sneath PHA, Sokal RR (1973) Numerical taxonomy. WH Freeman Press, San Francisco
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten TE, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald PC (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Takken FLW, Joosten MAHJ (2000) Plant resistance genes: their structure, function and evolution. Eur J Plant Pathol 106:699–713
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustalx windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian Y, Fan L, Thurau T, Jung C, Cai D (2004) The absence of TIR-type resistance gene analogues in the sugar beet (Beta vulgaris L.) genome. J Mol Evol 58:40–53
Traut TW (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide binding-sites. Eur J Biochem 222:9–19
Van Ooijen JW, Voorrips RE (2001) joinmap 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Vilanova S, Romero C, Abbott AG, Llácer G, Badenes ML (2003) An apricot (Prunus armeniaca L.) F2 progeny linkage map based on SSR and AFLP markers, mapping plum pox virus resistance and self-incompatibility traits. Theor Appl Genet 107:239–247
Vos P, Hogers V, Bleeker M, Reijans M, Van de Lee T, Hornes M, AA Frijters, Pot J, Pelman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Warren R, Henk A, Mowery P, Holub E, Innes RW (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPP5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10:1439–1452
Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B (1994) The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the Interleukin-1 receptor. Cell 78:1101–1115
Young ND (2000) The genetic architecture of resistance. Curr Opin Plant Biol 3:285–290
Acknowledgements
This research was supported by a grant from the Ministerio de Ciencia y Tecnología (AGL2001-1102-C02-02). The authors want to thank Donna Abernathy for her kind revision of this manuscript and José Martinez and Dolores Archelós for their technical contributions. J.M.S. was funded by a fellowship from the Ministerio de Ciencia y Tecnología of Spain. All of the experiments described in this paper comply with the current laws of Spain.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Möllers
Rights and permissions
About this article
Cite this article
Soriano, J.M., Vilanova, S., Romero, C. et al. Characterization and mapping of NBS-LRR resistance gene analogs in apricot (Prunus armeniaca L.). Theor Appl Genet 110, 980–989 (2005). https://doi.org/10.1007/s00122-005-1920-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-1920-0