Abstract
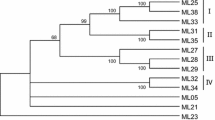
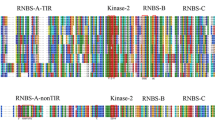
Toll and interleukin-1 receptor (TIR) and nonTIR nucleotide binding site–leucine rich repeat (NBS–LRR) resistance gene analogues (RGAs) were obtained from chestnut rose (Rosa roxburghii Tratt) by two PCR-based amplification strategies (direct amplification and overlap extension amplification) with degenerate primers designed to the conserved P-loop, kinase-2, and Gly-Leu-Pro-Leu (GLPL) motifs within the NBS domain of plant resistance gene (R gene) products. Thirty-four of 65 cloned PCR fragments contained a continuous open reading frame (ORF) and their predicted protein products showed homology to the NBS–LRR class R proteins in the GenBank database. These 34 predicted protein sequences exhibited a wide range (19.5–99.4%) of sequence identity among them and were classified into two distinct groups by phylogenetic analysis. The first group consisted of 23 sequences and seemed to belong to the nonTIR NBS–LRR RGAs, since they contained group specific motifs (RNBS-A-nonTIR motif) that are often present in the coiled-coil domain of the nonTIR NBS–LRR class R genes. The second group comprised 11 sequences that contained motifs found in the TIR domain of TIR NBS–LRR class R genes. Restriction fragment length polymorphic (RFLP) markers were developed from some of the RGAs and used for mapping powdery mildew resistance genes in chestnut rose. Three markers, RGA22C, RGA4A, and RGA7B, were identified to be linked to a resistance gene locus, designated CRPM1 for chestnut rose powdery mildew resistance 1, which accounted for 72% of the variation in powdery mildew resistance phenotype in an F1 segregating population. To our knowledge, this is the first report on isolation, phylogenetic analysis and potential utilization as genetic markers of RGAs in chestnut rose.








Similar content being viewed by others
References
Aarts MG, Hekkert BL, Holub EB, Beynon JL, Stiekema WJ, Pereira A (1998) Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant Microbe Interact 11:251–258
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programmes. Nucleic Acids Res 25:3389–3402
Ashfield T, Bocian A, Held D, Henk AD, Marek LF, Danesh D, Penuela S, Meksem K, Lightfoot DA, Young ND, Shoemaker RC, Innes RW (2003) Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS–LRR genes. Mol Plant Microbe Interact 16:817–826
Cana EF, Geffroy V, Macadre C, Creusot F, Imbert-Bollore P, Sevignac M, Langin T (2003) Characterization of expressed NBS–LRR resistance gene candidates from common bean. Theor Appl Genet 106:251–261
Collins NC, Webb CA, Seah S, Ellis JG, Hulber SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:968–978
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Deng Z, Huang S, Ling P, Chen C, Yu C, Weber CA, Moore GA, Gmitter FG (2000) Cloning and characterization of NBS–LRR class resistance-gene candidate sequences in citrus. Theor Appl Genet 101:814–822
Di Gaspero G, Cipriani G (2003) Nucleotide binding site/leucine-rich repeats, Pto-like and receptor-like kinases related to disease resistance in grapevine. Mol Gen Genomics 269:612–623
Donald TM, Pellerone F, Adam-Blondon AF, Bouquet A, Thomas MR, Dry IB (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor Appl Genet 101:301–308
Ellins JG, Lawrence GJ, Luck JE, Dodds N (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506
He LM, Du CG, Covaleda L, Xu ZY, Robinson AF, Yu JZ, Kohel RJ, Zhang HB (2004) Cloning, characterization, and evolution of the NBS–LRR-encoding resistance gene analogue family in polyploid cotton (Gossypium hirsutum L). Mol Plant Microbe Interact 17:1234–1241
Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59
Huang CC, Cui YY, Weng CR, Zabel P, Lindhout P (2000) Development of diagnostic PCR markers closely linked to the tomato powdery mildew resistance gene Ol-1 on chromosome 6 of tomato. Theor Appl Genet 101:918–924
Jones DA, Jones JDG (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24:90–167
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Aca Sci USA 93:11746–11750
Kumar S, Tamura K, Jakobsen I, Nei M (2000) Mega 2.0. http://www.megasoftware.net
Kunoh H (1995) Host-parasite specificity in powdery mildews. In:Singh US, Kohmoto K, Singh RP (eds) Pathogenesis and host specificity in plant diseases: histopathological, biochemical, genetic and molecular bases, vol. 2 Eukaryotes. Elsevier, Oxford, pp 239–250
Lambert P, Hagen LS, Arus P, Audergon JM (2004) Genetic linkage maps of two apricot cultivars (Prunus armeniaca L.) compared with the almond Texas×peach Earlygold reference map for Prunus. Theor Appl Genet 108:1120–1130
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefer P (1998) Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA 95:370–375
Linde M, Mattiesch L, Debener T (2004) Rpp1, a dominant gene providing race-specific resistance to rose powdery mildew (Podosphaera pannosa): molecular mapping, SCAR development and confirmation of disease resistance data. Theor Appl Genet 109:1261–1266
Liu L, Kloepper JW, Tuzun S (1996) Induction of systemic induced resistance in cucumber against Fusarium wilt by plant growth-promoting rhizobacteria. Phytopathology 85:695–698
Ma YX, Zhu Y, Wang CF (1997) The aging retarding effect of ‘Long-Life CiLi’. Mech Ageing Dev 96:171–189
McDowell JM, Dhandaydham M, Long TA, Aatrs MG, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10:1861–1874
Meer J, Cudmore R, Manly KF (2004) Map Manager QTX. http://www.mapmanager.org/mmQTX.html
Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS–LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Michelmore RW, Paran l, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Monosi B, Wisser R J, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Noir S, Combes M-C, Anthony F, Lashermes P (2001) Origin, diversity and evolution of NBS-type disease-resistance gene homologues in coffee trees (Coffea L.). Mol Gen Genomics 265:654–662
Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS–LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50:203–213
Penuela S, Danesh D, Young ND (2002) Targeted isolation, sequence analysis, and physical mapping of nonTIR NBS–LRR genes in soybean. Theor Appl Genet 104:261–272
Radwan O, Bouzidi MF, Nicolas P, Mouzeyar S (2004) Development of PCR markers of the PI5/PI8 locus for resistance to Plasmopara halstedii in sunflower, Helianthus annuus L. from complete CC-NBS–LRR sequences. Theor Appl Genet 109:176–185
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbour Laboratory, New York, pp 19–22
Song WY, Pi LY, Wang GL, Gardner J. Olsten T, Ronald PC (1997) Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9:1279–1287
Speulman E, Bouchez D, Holub EB, Beynon JL (1998) Disease resistance gene homologs correlate with disease resistance loci of Arabidopsis thaliana. Plant J 14:467–474
Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400:667–671
Tameling WIL, Elzinga SDJ, Darmin PS, Vossen JH, Takken FLW, Harling MA, Cornelissen B (2002) The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14:2929–2939
Tanhuanpaa P (2004) Identification and mapping of resistance gene analogs and a white rust resistance locus in Brassica rapa ssp. Oleifera. Theor Appl Genet 108:1039–1046
Teruel MN, Meyer T (2000) Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell 2:181–184
Tian Y, Fan L, Thurau T, Jung C, Cai D (2004) The absence of TIR-type resistance gene analogues in the sugar beet (Beta vulgaris L.) genome. J Mol Evol 58:40–53
Van der Biezen EA, Sun J, Coleman MJ, Bibb MJ, Jones JDG (2000) Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc Natl Acad Sci USA 97:3747–3752
Wen XP, Pang XM, Deng XX (2004) A comparative study of morphological, RAPD, AFLP approaches to characterize relationships of Rosa roxburghii Tratt and its relatives. J Hortic Sci Biotechnol 79:189–196
Wen XP, Deng XX (2005a) Micropropagation of chestnut rose (Rosa roxburghii Tratt) and genetic stability assessment of the in vitro plants using RAPD and AFLP markers. J Hortic Sci Biotechnol 80:54–60
Wen XP, Xu Q, Fan W, Deng X (2005b) A preliminary study on inheritance tendency of the resistance to powdery mildew using F1 progenies of chestnut rose (Rosa roxburghii Tratt). Acta Horticulturae Sinica 320:304–306
Xiao SY, Ellwood S, Callis O, Patrick E, Li TX, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291:118–120
Xu Q, Wen XP, Deng XX (2004) A simple protocol for isolation of genomic DNA from chestnut rose (Rosa roxburghii Tratt) for PCR and RFLP analysis. Plant Mol Biol Reptr 22:301–302
Yu YG, Buss GR, Maroof MA (1996) Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Zhu H, Cannon SB, Young ND, Cook DR (2002) Phylogeny and genomic organization of the TIR and nonTIR NBS–LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact 15:529–539
Acknowledgements
This project was supported by the National Natural Science Foundation of China (NSFC) (Nos.30260070 30123001 and 30471201), the 863 Project of China, and Natural Science Foundation of Guizhou Province. The authors are grateful to Dr. S. Xiao from Center for Biosystems Research of University of Maryland Biotechnology institute, Dr. Z. Deng from Gulf Coast Research and Education Center of University of Florida, and Dr. W.W. Guo from our laboratory for critical review and helpful advice, and to Prof. K. Watanabe from Plant Genetic Diversity Gene Research Center of Tsukuba University (Japan) for his reading the manuscript. Our appreciations also extend to Mr. Y.Z. Liu, J.K. Song, X.D. Cai and S.H. Zeng in our laboratory for their assistance with the experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Nybom
Rights and permissions
About this article
Cite this article
Xu, Q., Wen, X. & Deng, X. Isolation of TIR and nonTIR NBS–LRR resistance gene analogues and identification of molecular markers linked to a powdery mildew resistance locus in chestnut rose (Rosa roxburghii Tratt). Theor Appl Genet 111, 819–830 (2005). https://doi.org/10.1007/s00122-005-0002-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0002-7