Abstract
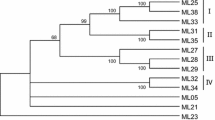
Degenerate oligonucleotide primers, designed based on conserved regions of Nucleotide Binding Site (NBS) domains from previously cloned plant resistance genes, were used to isolate Resistance Gene Analogues (RGAs) from wild and cultivated strawberries. Seven distinct families of RGAs of the NBS-LRR type were identified from two related wild species, Fragaria vesca and F. chiloensis, and six different Fragaria × ananassa cultivars. With one exception (GAV-3), the deduced amino acid sequences of strawberry RGAs showed strong similarity to TIR (Toll Interleukin I Receptor)-type R genes from Arabidopsis, tobacco and flax, suggesting the existence of common ancestors. GAV-3 seemed to be more closely related to the non-TIR type. Further studies showed that the recombination level and the ratio of non-synonymous to synonymous substitutions within families were low. These data suggest that NBS-encoding sequences of RGAs in strawberry are subject to a gradual accumulation of mutations leading to purifying selection, rather than to a diversifying process. The present paper is the first report on RGAs in strawberry.



Similar content being viewed by others
References
Aarts MG, te Lintel Hekkert B, Holub BT, Beynon JL, Stiekema WJ, Pereira A (1998) Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant-Microbe Interact 11:251–258
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif., pp 28–36
Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP (1997) Signaling in plant-microbe interactions. Science 276:726–733
Bent AF (1996) Plant disease resistance gene: function meets structure. Plant Cell 8:1757–1771
Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117–127
Castagnaro A, Maraña C, Carbonero P, García-Olmedo E (1992) Extreme divergence of a novel wheat thionin generated by a mutational burst specifically affecting the mature protein domain of the precursor. J Mol Biol 224:1003–1009
Cordero JC, Skinner DZ (2002) Isolation from alfalfa of resistance gene analogues containing nucleotide binding sites. Theor Appl Genet 104:1283–1289
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Ellis J, Dodds P, Pryor T (2000) Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol 3:278–284
Felsenstein J (1985) Confidence limits on phylogenesis: an approach using the bootstrap. Evolution 39:783–791
Fluhr R (2001) Sentinels of disease. Plant resistance genes. Plant Physiol 127:1367–1374
Galletta GJ, Smith BJ, Gupton CL (1993) Strawberry parent clones US70, US159, US292, and US438 resistant to anthracnose crown rot. Hortscience 28:1055–1056
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Haymes KM, Henken B, Davis TM, van de Weg WE (1997) Identification of RAPD markers linked to a Phytophthora fragariae resistance gene ( Rpf1) in the cultivated strawberry. Theor Appl Genet 94:1097–1101
Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana DNA by combining local and global sequence information. Nucleic Acids Research 24:3439–3452
Hofman K, Bucher P, Falquet L, Bairoch A (1999) The PROSITE database, its status in 1999. Nucleic Acids Res 27:215–219
Huettel B, Santra D, Muehlbauer FJ, Kahl G (2002) Resistance gene analogues of chickpea ( Cicer arietinum L.): isolation, genetic mapping and association with a Fusarium resistance gene cluster. Theor Appl Genet 105:479-490
Hulbert S H, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Joyeux A, Fortin MG, Mayerhofer R, Good AG (1999) Genetic mapping of plant disease resistance gene homologues using a minimal Brassica napus L. population. Genome 42:735–743
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide seqeunce. J Mol Biol 16:111–120
Kobe B, Deisenhofer J (1996) Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol 264:1028–1043
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244–1245
Lee SY, Seo JS, Rodriguez-Lanetty M, Lee DH (2003) Comparative analysis of superfamilies of NBS-encoding disease resistance gene analogs in cultivated and wild apple species. Mol Genet Genomics 269:101–108
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Lerceteau-Kohler E, Roudeillac P, Markocic M, Guerin G, Praud K, Denoyes-Rohan B (2002) The use of molecular markers for durable resistance breeding in the cultivated strawberry ( Fragaria x ananassa). Acta Hort 567:615–618
Martin GB (1999) Functional analysis of plant disease resistance genes and their downstream effectors. Curr Opin Plant Biol 2:273–279
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11:2099–2112
Noir S, Combes MC, Anthony F, Lashermes P (2001) Origin, diversity and evolution of NBS-type disease-resistance gene homologues in coffee trees ( Coffea L.). Mol Genet Genomics 265:654–662
Pan Q, Liu YS, Budai-Hadrian O, M S, Carmel-Goren L, Zamir D, Fluhr R (2000) Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155:309–322
Parker JE, Coleman MJ, Szabo V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JDG (1997) The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and interleukin-1 receptors with N and L6. Plant Cell 9:879–894
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Saraste M, Sibbald PR, Wittinghofer A (1990) The P-loop a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci 15:430–434
Sawyer S (1989) Statistical tests for detecting gene conversion. Mol Biol Evol 6:526–538
Shen KA, Meyers BC, Nurul Islam-Faridi M, Chin DB, Stelly DM, Michelmore RW (1998) Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol Plant Microbe Interact 11:815–823
Speulman E, Bouchez D, Holub EB, Beynon JL (1998) Disease resistance gene homologs correlate with disease resistance loci of Arabidopsis thaliana. Plant J 14:467–474
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian Y, Fan L, Thurau, T, Jung C, Cai D (2004) The absence of TIR-type resistance gene analogues in the sugar beet ( Beta vulgaris L.) genome. J Mol Evol 58:40–53
Traut TW (1994) The functions and consensus motifs of nine types of peptide segments that form di§erent types of nucleotide-binding sites. Eur J Biochem 222:9–19
Weising K, Atkinson RG, Gardner RC (1995) Genomic fingerprinting by microsatellite-primed PCR: a critical evaluation. PCR Methods Appl 4:249–255
Yu YG, Buss GR, Maroof MA (1996) Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Acknowledgements
This paper was partially supported by grants PICT-7227, PICT-7229 from FONCYT and 26/D209 from CIUNT. Degenerate PCR primers were kindly provided by Dr. Fernando García-Arenal from ETSIA-Universidad Politécnica de Madrid, Spain. This work was carried out in compliance with the current laws regulating genetic experimentation in Argentina. GMZ is a fellow from CONICET and APC and JCDR are members of CONICET
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M.-A. Grandbastien
Rights and permissions
About this article
Cite this article
Martínez Zamora, M.G., Castagnaro, A.P. & Díaz Ricci, J.C. Isolation and diversity analysis of resistance gene analogues (RGAs) from cultivated and wild strawberries. Mol Genet Genomics 272, 480–487 (2004). https://doi.org/10.1007/s00438-004-1079-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-004-1079-4