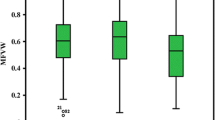
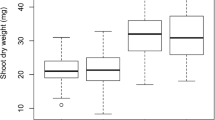
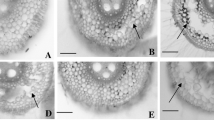
Abstract
Main conclusion
37 unconditional QTLs, 51 conditional QTLs and considerable epistatic QTLs were detected for waterlogging tolerance, and six favourable combinations were selected accelerating the possible application of MAS in chrysanthemum breeding.
Chrysanthemum is seriously impacted by soil waterlogging. To determine the genetic characteristics of waterlogging tolerance (WAT) in chrysanthemum, a population of 162 F1 lines was used to construct a genetic map to identify the dynamic and epistatic quantitative trait loci (QTLs) for four WAT traits: wilting index (WI), dead leaf ratio (DLR), chlorosis score (Score) and membership function value of waterlogging (MFVW). The h 2B for the WAT traits ranged from 0.49 to 0.64, and transgressive segregation was observed in both directions. A total of 37 unconditional consensus QTLs with 5.81–18.21% phenotypic variation explanation (PVE) and 51 conditional consensus QTLs with 5.90–24.56% PVE were detected. Interestingly, three unconditional consensus QTLs were consistently identified across different stages, whereas no conditional consensus QTLs were consistently expressed. In addition, considerable epistatic QTLs, all with PVE values ranging from 0.01 to 8.87%, were detected by a joint analysis of WAT phenotypes. These results illustrated that the QTLs (genes) controlling WAT were environmentally dependent and selectively expressed at different times and indicated that both additive and epistatic effects underlie the inheritance of WAT in chrysanthemum. The findings of the current study provide insights into the complex genetic architecture of WAT, and the identification of favourable alleles represents an important step towards the application of molecular marker-assisted selection (MAS) and QTL pyramiding in chrysanthemum WAT breeding programmes.







Similar content being viewed by others
Abbreviations
- DLR:
-
Dead leaf ratio
- gSSR:
-
Genomic simple sequence repeat
- LG:
-
Linkage group
- LOD:
-
Logarithm of odds
- MAS:
-
Marker assisted selection
- MB:
-
Monalisa cv
- MFVW:
-
Membership function value of waterlogging
- PVE:
-
Phenotypic variance explained
- QTL:
-
Quantitative trait loci
- Score:
-
Chlorosis score
- SRAP:
-
Sequence-related amplified polymorphism
- WAT:
-
Waterlogging tolerance
- WI:
-
Wilting index
- XF:
-
Nannong Xuefeng cv
- XM:
-
XF and MB population
References
Anderson NO, Ascher PD (2000) Fertility changes in inbred families of self-incompatible chrysanthemums (Dendranthema grandiflora). J Am Soc Hortic Sci 125(5):619–625
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20(14):2324–2326
Atchley WR, Zhu J (1997) Developmental quantitative genetics, conditional epigenetic variability and growth in mice. Genetics 147(2):765–776
Bian Y, Gu X, Sun D, Wang Y, Yin Z, Deng D, Wang Y, Li G (2015) Mapping dynamic QTL of stalk sugar content at different growth stages in maize. Euphytica 205(1):85–94
Boeven PHG, Longin CFH, Leiser WL, Kollers S, Ebmeyer E, Würschum T (2016) Genetic architecture of male floral traits required for hybrid wheat breeding. Theor Appl Genet 129(12):2343–2357
Cai C, Cheng FY, Wu J, Zhong Y, Liu G (2015) The first high-density genetic map construction in tree peony (Paeonia Sect. Moutan) using genotyping by specific-locus amplified fragment sequencing. PLoS One 10(5):e0128584
Chakravarti A, Lasher L, Reefer J (1991) A maximum likelihood method for estimating genome length using geneticlinkage data. Genetics 128(1):175–182
Chong X, Zhang F, Wu Y, Yang X, Zhao N, Wang H, Guan Z, Fang W, Chen F (2016) A SNP-enabled assessment of genetic diversity, evolutionary relationships and the identification of candidate genes in chrysanthemum. Genome Biol Evol 8(12):3661–3671
Cornelious B, Chen P, Chen Y, De Leon N, Shannon JG, Wang D (2005) Identification of QTLs underlying water-logging tolerance in soybean. Mol Breed 16(2):103–112
Desnoues E, Baldazzi V, Génard M, Mauroux JB, Lambert P, Confolent C, Quilot-Turion B (2016) Dynamic QTLs for sugars and enzyme activities provide an overview of genetic control of sugar metabolism during peach fruit development. J Exp Bot 67(11):3419–3431
Falconer DS, Machay TFC (1996) Introduction to quantitative genetics, 4th edn. Longmans Green, Harlow, pp 1–90
Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155(1):463–473
Guo H, Ding W, Chen J, Chen X, Zheng Y, Wang Z, Liu J (2014) Genetic linkage map construction and QTL mapping of salt tolerance traits in zoysiagrass (Zoysia japonica). PLoS One 9(9):e107249
Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, Matsuoka A, Mori H, Ashikari M (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460(7258):1026–1030
Hund A, Fracheboud Y, Soldati A, Frascaroli E, Salvi S, Stamp P (2004) QTL controlling root and shoot traits of maize seedlings under cold stress. Theor Appl Genet 109(3):618–629
Kadam S, Abril A, Dhanapal AP, Koester RP, Vermerris W, Jose S, Fritschi FB (2017) Characterization and regulation of aquaporin genes of sorghum [Sorghum bicolor (L) Moench] in response to waterlogging stress. Front Plant Sci 8:862
Kim C, Zhang D, Auckland SA, Rainville LK, Jakob K, Kronmiller B, Sacks EJ, Deuter M, Paterson AH (2012) SSR-based genetic maps of Miscanthus sinensis and M. sacchariflorus, and their comparison to sorghum. Theor Appl Genet 124(7):1325–1338
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12(1):172–175
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103(2):455–461
Li J, Lindqvist-Kreuze H, Tian Z, Liu J, Song B, Landeo J, Portal L, Gastelo M, Frisancho J, Sanchez L (2012) Conditional QTL underlying resistance to late blight in a diploid potato population. Theor Appl Genet 124(7):1339–1350
Li Z, Mei S, Mei Z, Liu X, Fu T, Zhou G, Tu J (2014) Mapping of QTL associated with waterlogging tolerance and drought resistance during the seedling stage in oilseed rape (Brassica napus). Euphytica 197(3):341–353
Liang Q, Li P, Hu C, Hua H, Li Z, Rong Y, Wang K, Hua J (2014) Dynamic QTL and epistasis analysis on seedling root traits in upland cotton. J Genet 93(1):63–78
Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62(2):302–315
Liu L, Lai Y, Cheng J, Wang L, Du W, Wang Z, Zhang H (2014) Dynamic quantitative trait locus analysis of seed vigor at three maturity stages in rice. PLoS One 9(12):e115732
McCouch SR (2008) Gene nomenclature system for rice. Rice 1(1):72–84
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4326
Oliveira KM, Pinto LR, Marconi TG, Margarido GR, Pastina MM, Teixeira LHM, Figueira AV, Ulian EC, Garcia AAF, Souza AP (2007) Functional integrated genetic linkage map based on EST-markers for a sugarcane (Saccharum spp.) commercial cross. Mol Breed 20(3):189–208
Osman KA, Tang B, Wang Y, Chen J, Yu F, Li L, Han X, Zhang Z, Yan J, Zheng Y (2013) Dynamic QTL analysis and candidate gene mapping for waterlogging tolerance at maize seedling stage. PLoS One 8(11):e79305
Peng H, Zhang F, Jiang J, Chen S, Fang W, Guan Z, Chen F (2015) Identification of quantitative trait loci for branching traits of spray cut chrysanthemum. Euphytica 202(3):385–392
Qi X, Xu X, Lin X, Zhang W, Chen X (2012) Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics 99(3):160–168
Qiu F, Zheng Y, Zhang Z, Xu S (2007) Mapping of QTL associated with waterlogging tolerance during the seedling stage in maize. Ann Bot 99(6):1067–1081
Rahman H, Pekic S, Lazic-Jancic V, Quarrie S, Shah S, Pervez A, Shah M (2011) Molecular mapping of quantitative trait loci for drought tolerance in maize plants. Genet Mol Res 10(2):889–901
Ravi K, Vadez V, Isobe S, Mir R, Guo Y, Nigam S, Gowda M, Radhakrishnan T, Bertioli D, Knapp S (2011) Identification of several small main-effect QTLs and a large number of epistatic QTLs for drought tolerance related traits in groundnut (Arachis hypogaea L.). Theor Appl Genet 122(6):1119–1132
Shang L, Cai S, Ma L, Wang Y, Abduweli A, Wang M, Wang X, Liang Q, Hua J (2016) Seedling root QTLs analysis on dynamic development and upon nitrogen deficiency stress in upland cotton. Euphytica 207(3):645–663
Shao H, Burrage LC, Sinasac DS, Hill AE, Ernest SR, O’Brien W, Courtland HM, Jepsen KJ, Kirby A, Kulbokas EJ, Daly MJ, Broman KW, Lander ES, Nadeau JH (2008) Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc Natl Acad Sci USA 105(50):19910–19914
Strigens A, Freitag NM, Gilbert X, Grieder C, Riedelsheimer C, Schrag TA, Messmer R, Melchinger AE (2013) Association mapping for chilling tolerance in elite flint and dent maize inbred lines evaluated in growth chamber and field experiments. Plant Cell Environ 36(10):1871–1887
Su J, Zhang F, Li P, Guan Z, Fang W, Chen F (2016) Genetic variation and association mapping of waterlogging tolerance in chrysanthemum. Planta 244(6):1241–1252
Su J, Zhang F, Yang X, Feng Y, Yang X, Wu Y, Guan Z, Fang W, Chen F (2017) Combining ability, heterosis, genetic distance and their intercorrelations for waterlogging tolerance traits in chrysanthemum. Euphytica 213(2):42. https://doi.org/10.1007/s10681-017-1837-0
Teixeira da Silva JA, Shinoyama H, Aida R, Matsushita Y, Raj SK, Chen F (2013) Chrysanthemum biotechnology: Quo vadis? Crit Rev Plant Sci 32(1):21–52
Thirunavukkarasu N, Hossain F, Mohan S, Shiriga K, Mittal S, Sharma R, Singh RK, Gupta HS (2013) Genome-wide expression of transcriptomes and their co-expression pattern in subtropical maize (Zea mays L.) under waterlogging stress. PLoS One 8(8):e70433
Ungerer MC, Halldorsdottir SS, Modliszewski JL, Mackay TF, Purugganan MD (2002) Quantitative trait loci for inflorescence development in Arabidopsis thaliana. Genetics 160(3):1133–1151
Van Ooijen J (2006) JoinMap 4: Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Voorrips R (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Vukosavljev M, Arens P, Voorrips RE, van’t Westende WP, Esselink G, Bourke PM, Cox P, Van De Weg WE, Visser RG, Maliepaard C (2016) High-density SNP-based genetic maps for the parents of an outcrossed and a selfed tetraploid garden rose cross, inferred from admixed progeny using the 68 k rose SNP array. Hortic Res 3:16052
Wang D, Zhu J, Li Z, Paterson A (1999) Mapping QTLs with epistatic effects and QTL × environment interactions by mixed linear model approaches. Theor Appl Genet 99(77):1255–1264
Wang S, Basten C, Zeng Z (2007) Windows QTL cartographer 25 department of statistics. North Carolina State University, Raleigh
Wang Z, Wu X, Ren Q, Chang X, Li R, Jing R (2010) QTL mapping for developmental behavior of plant height in wheat (Triticum aestivum L.). Euphytica 174(3):447–458
Wang Z, Cheng J, Chen Z, Huang J, Bao Y, Wang J, Zhang H (2012) Identification of QTLs with main, epistatic and QTL × environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor Appl Genet 125(4):807–815
Wang H, Jiang J, Chen S, Qi X, Peng H, Li P, Song A, Guan Z, Fang W, Liao Y (2013) Next-generation sequencing of the Chrysanthemum nankingense (Asteraceae) transcriptome permits large-scale unigene assembly and SSR marker discovery. PLoS One 8(4):e62293
Wang C, Zhang F, Guan Z, Chen S, Jiang J, Fang W, Chen F (2014a) Inheritance and molecular markers for aphid (Macrosiphoniella sanbourni) resistance in chrysanthemum (Chrysanthemum morifolium Ramat.). Sci Hortic 180:220–226
Wang L, Cheng J, Lai Y, Du W, Huang X, Wang Z, Zhang H (2014b) Identification of QTLs with additive, epistatic and QTL × development interaction effects for seed dormancy in rice. Planta 239(2):411–420
Wang X, Wang H, Long Y, Liu L, Zhao Y, Tian J, Zhao W, Li B, Chen L, Chao H (2015) Dynamic and comparative QTL analysis for plant height in different developmental stages of Brassica napus L. Theor Appl Genet 128(6):1175–1192
Weng Y, Colle M, Wang Y, Yang L, Rubinstein M, Sherman A, Ophir R, Grumet R (2015) QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor Appl Genet 128(9):1747–1763
Wu WR, Li WM, Tang DZ, Lu HR, Worland A (1999) Time-related mapping of quantitative trait loci underlying tiller number in rice. Genetics 151(1):297–303
Wu P, Liao C, Hu B, Yi K, Jin W, Ni J, He C (2000) QTLs and epistasis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Theor Appl Genet 100(8):1295–1303
Würschum T, Liu W, Busemeyer L, Tucker MR, Reif JC, Weissmann EA, Hahn V, Ruckelshausen A, Maurer HP (2014) Mapping dynamic QTL for plant height in triticale. BMC Genet 15(1):59
Xu K, Xia X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AI, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442(7103):705–708
Yaboah MA, Chen X, Cheng RF, Alfandi M, Liang G, Gu M (2008) Mapping quantitative trait loci for waterlogging tolerance in cucumber using SRAP and ISSR markers. Biotechnology 7(2):157–167
Yang J, Zhu J (2005) Methods for predicting superior genotypes under multiple environments based on QTL effects. Theor Appl Genet 110(7):1268–1274
Yang J, Zhu J, Williams RW (2007) Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23(12):1527–1536
Yang X, Guo Y, Yan J, Zhang J, Song T, Rocheford T, Li J (2010) Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor Appl Genet 120(3):665–678
Yin D, Guan Z, Chen S, Chen F (2009a) Establishment of evaluation system for waterlogging tolerance and identification of waterlogging tolerance in Chrysanthemum morifolium and its related genera plants. J Plant Genet Res 10(3):399–404
Yin D, Chen S, Chen F, Guan Z, Fang W (2009b) Morphological and physiological responses of two chrysanthemum cultivars differing in their tolerance to waterlogging. Environ Exp Bot 67(1):87–93
Yin D, Ni D, Song L, Zhang Z (2013) Isolation of an alcohol dehydrogenase cDNA from and characterization of its expression in chrysanthemum under waterlogging. Plant Sci 212(7):48–54
Yu S, Li J, Xu C, Tan Y, Gao Y, Li X, Zhang Q, Maroof MS (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 94(17):9226–9231
Zanke CD, Rodemann B, Ling J, Muqaddasi QH, Plieske J, Polley A, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Zschäckel T, Ganal MW, Roder MS (2017) Genome-wide association mapping of resistance to eyespot disease (Pseudocercosporella herpotrichoides) in European winter wheat (Triticum aestivum L.) and fine-mapping of Pch1. Theor Appl Genet 130(3):505–514
Zeng ZB (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90(23):10972–10976
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136(4):1457–1468
Zhang F, Chen S, Chen F, Fang W, Li F (2010) A preliminary genetic linkage map of chrysanthemum (Chrysanthemum morifolium) cultivars using RAPD. ISSR and AFLP markers. Sci Hortic 125(3):422–428
Zhang F, Chen S, Chen F, Fang W, Chen Y, Li F (2011) SRAP-based mapping and QTL detection for inflorescence-related traits in chrysanthemum (Dendranthema morifolium). Mol Breed 27(1):11–23
Zhang F, Jiang J, Chen S, Chen F, Fang W (2012a) Detection of quantitative trait loci for leaf traits in chrysanthemum. J Hortic Sci Biotech 87(6):613–618
Zhang F, Jiang J, Chen S, Chen F, Fang W (2012b) Mapping single-locus and epistatic quantitative trait loci for plant architectural traits in chrysanthemum. Mol Breed 30(2):1027–1036
Zhang F, Chen S, Jiang J, Guan Z, Fang W, Chen F (2013) Genetic mapping of quantitative trait loci underlying flowering time in chrysanthemum (Chrysanthemum morifolium). PLoS One 8(12):e83023
Zhang J, Zhang Q, Cheng T, Yang W, Pan H, Zhong J, Huang L, Liu E (2015) High-density genetic map construction and identification of a locus controlling weeping trait in an ornamental woody plant (Prunus mume Sieb. et Zucc). DNA Res 22(3):183–191
Zhang X, Fan Y, Shabala S, Koutoulis A, Shabala L, Johnson P, Hu H, Zhou M (2017) A new major-effect QTL for waterlogging tolerance in wild barley (H. spontaneum). Theor Appl Genet 130(8):1559–1568
Zhu J (1995) Analysis of conditional genetic effects and variance components in developmental genetics. Genetics 141(4):1633–1639
Acknowledgements
This work was financially supported by the National Science Fund for Distinguished Young Scholars (31425022), Key Projects of National Natural Science Foundation of China (31730081), Fund for Independent Innovation of Agricultural Sciences in Jiangsu Province [CX (16) 1025], Special Fund for Agroscientific Research in the Public Interest (201403039), “948” Project of Ministry of Agriculture (2016-X18), and Fundamental Research Funds for the Central Universities (KYRC201601, KYCYL201501).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2017_2833_MOESM1_ESM.tif
Fig. S1 The daily mean temperature (a) and relative humidity (b) at the four measured stages in the two environments (TIFF 2847 kb)
425_2017_2833_MOESM2_ESM.tif
Fig. S2 A genetic linkage map for chrysanthemum XF based on testcross markers at an LOD threshold of 4.0. The cumulative genetic distances (cM) are indicated on the left side of each LG, and the names of the markers are on the right side. Markers with red colour are gSSR, and the prefix * or ** indicates a segregation distortion at P < 0.05 and P < 0.01, respectively (TIFF 10204 kb)
425_2017_2833_MOESM3_ESM.tif
Fig. S3 A genetic linkage map for chrysanthemum MB based on testcross markers at an LOD threshold of 4.0. The cumulative genetic distances (cM) are indicated on the left side of each LG, and the names of the markers are on the right side. Markers with red colour are gSSR, and the prefix * or ** indicates a segregation distortion at P < 0.05 and P < 0.01, respectively (TIFF 13472 kb)
425_2017_2833_MOESM4_ESM.tif
Fig. S4 MFVW values of XF, MB, and the seven selected F1 lines at four stages in environments E1 (a) and E2 (b) (TIFF 2821 kb)
Rights and permissions
About this article
Cite this article
Su, J., Yang, X., Zhang, F. et al. Dynamic and epistatic QTL mapping reveals the complex genetic architecture of waterlogging tolerance in chrysanthemum. Planta 247, 899–924 (2018). https://doi.org/10.1007/s00425-017-2833-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2833-2