Abstract
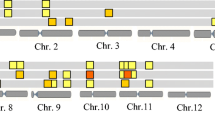
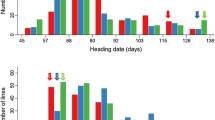
Seed dormancy (SD) is an important agronomic trait affecting crop yield and quality. In this study, one rice population of recombinant inbred lines (RILs) was used to determine the genetic characteristics of SD at the early (4 weeks after heading), middle (5 weeks after heading) and late (6 weeks after heading) development stages. The level of SD decreased with the process of seed development, and the SD was significantly affected by the heading date (HD) and temperature at the early and middle development stages. A total of eight additive quantitative trait loci (QTLs) for SD were identified at three development stages, and more QTLs were expressed at the early and late development stages. Among them, four, one and three additive QTLs were identified at the early, middle and late development stages, respectively. Epistatic QTLs and QTL-by-development interactions were detected by the joint analysis of multi-development phenotypic values, and one additive and two epistatic QTLs were identified. The phenotypic variation of SD explained by each additive, epistatic QTL and QTL × development interaction ranged from 8.0 to 13.5 %, 0.7 to 3.9 % and 1.3 to 2.8 %, respectively. One major QTL qSD7.1 for SD was tightly linked to the major QTL qHD7.4 for HD, which might be applied to reveal the relationship of SD and HD. By comparing chromosomal positions of these additive QTLs with those previously identified, five additive QTLs qSD1.1, qSD2.1, qSD2.2, qSD4.1 and qSD4.2 might represent novel genes. The best three cross combinations for the development of RIL populations were predicted to improve SD. The selected RILs and the identified QTLs might be applicable to improve the rice pre-harvest sprouting tolerance by the marker-assisted selection approach.





Similar content being viewed by others
Abbreviations
- GP:
-
Germination percentage
- HD:
-
Heading date
- MAS:
-
Marker-assisted selection
- PHS:
-
Pre-harvest sprouting
- QTLs:
-
Quantitative trait loci
- RILs:
-
Recombinant inbred lines
- SD:
-
Seed dormancy
- SSR:
-
Simple sequence repeats
References
Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103:17042–17047
Biddulph TB, Plummer JA, Setter TL, Mares DJ (2007) Influence of high temperature and terminal moisture stress on dormancy in wheat (Triticum aestivum L.). Field Crops Res 103:139–153
Buraas T, Skinnes H (1985) Development of seed dormancy in barley, wheat and triticale under controlled conditions. Acta Agric Scand 35:233–244
Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor Appl Genet 95:553–567
Chen CX, Cai SB, Bai GH (2008) A major QTL controlling seed dormancy and pre-harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Mol Breed 21:351–358
Chiang GCK, Bartsch M, Barua D, Nakabayashi K, Debieu M, Kronholm I, Koornneef M, Soppe WJJ, Donohue K, De Meaux J (2011) DOG1 expression is predicted by the seed maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol Ecol 20:3336–3349
Dellaporta SL, Wood T, Hicks TB (1983) A plant DNA mini preparation: version II. Plant Mol Bio Rep 1:19–21
Dong Y, Tsuzukia E, Kamiuntena H, Teraoa H, Lina D, Matsuoa M, Zheng Y (2003) Identification of quantitative trait loci associated with pre-harvest sprouting resistance in rice (Oryza sativa L.). Field Crops Res 81:133–139
Fenner M (1991) The effects of the parent environment on seed germinability. Seed Sci Res 1:75–84
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415
Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE (2011) Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA 108:20236–20241
Gao W, Clancy JA, Han F, Prada D, Kleinhofs A, Ullrich SE (2003) Molecular dissection of a dormancy QTL region near the chromosome 7 (5H) L telomere in barley. Theor Appl Genet 107:552–559
Gao FY, Ren GJ, Lu XJ, Sun SX, Li HJ, Gao YM, Lou H, Yan WG, Zhang YZ (2008) QTL analysis for resistance to preharvest sprouting in rice (Oryza sativa). Plant Breed 127:268–273
Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35:1769–1786
Gu XY, Kianian SF, Foley ME (2004) Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa L.). Genetics 166:1503–1516
Gu XY, Liu T, Feng J, Suttle JC, Gibbons J (2010) The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice. Plant Mol Biol 73:97–104
Gualano NA, Benech-Arnold RL (2009) Predicting pre-harvest sprouting susceptibility in barley: looking for ‘‘sensitivity windows’’ to temperature throughout grain filling in various commercial cultivars. Field Crops Res 114:35–44
Guo L, Zhu L, Xu Y, Zeng D, Wu P, Qian Q (2004) QTL analysis of seed dormancy in rice (Oryza sativa L.). Euphytica 140:155–162
Hillhorst HWM (1995) A critical update on seed dormancy I. Primary dormancy. Seed Sci Res 5:61–73
Hori K, Sugimoto K, Nonoue Y, Ono N, Matsubara K, Yamanouchi U, Abe A, Takeuchi Y, Yano M (2010) Detection of quantitative trait loci controlling pre-harvest sprouting resistance by using backcrossed populations of japonica rice cultivars. Theor Appl Genet 120:1547–1557
Kendall SL, Hellwege A, Marriot P, Whalley C, Graham IA, Penfield S (2011) Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23:2568–2580
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Li H, Ribaut JM, Li Z, Wang J (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260
Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96:997–1003
Liu S, Bai G (2010) Dissection and fine mapping of a major QTL for preharvest sprouting resistance in white wheat Rio Blanco. Theor Appl Genet 121:1395–1404
Lu B, Xie K, Yang C, Wang S, Liu X, Zhang L, Jiang L, Wan J (2011) Mapping two major effect grain dormancy QTL in rice. Mol Breeding 28:453–462
Martinez-Andujar C, Ordiz MI, Huang Z, Nonogaki M, Beachy RN, Nonogaki H (2011) Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc Natl Acad Sci USA 108:17225–17229
McCouch SR, CGSNL (Committee on Gene Symbolization, Nomenclature, Linkage, Rice Genetics Cooperative) (2008) Gene nomenclature system for rice. Rice 1:72–84
McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu BY, Maghirang R, Li ZK, Xing YZ, Zhang QF, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2,240 new SSR markers for rice (Oryza sativa L.). DNA Res 6:199–207
Miura K, Araki H (1999) Effect of temperature during the ripening period on the induction of secondary dormancy in rice seeds (Oryza sativa L.). Breed Sci 49:7–10
Miyoshi K, Sato T (1997a) The effects of ethanol on the germination of seeds of japonica and indica rice (Oryza sativa L.) under anaerobic and aerobic conditions. Ann Bot 79:391–395
Miyoshi K, Sato T (1997b) Removal of the pericarp and tests of seeds of japonica and indica rice (Oryza sativa) at various oxygen concentrations has opposite effects on germination. Physiol Plant 99:1–6
Miyoshi K, Sato T, Takahashi N (1996) Differences in the effects of dehusking during formation of seeds on the germination of seeds of indica and japonica rice (Oryza sativa L.). Ann Bot 77:599–604
Nicholls PB (1982) Influence of temperature during grain growth and ripening of barley on the subsequent response to exogenous gibberellic acid. Aust J Plant Physiol 9:373–383
Niu Y, Xu Y, Liu XF, Yang SX, Wei SP, Xie FT, Zhang YM (2013) Association mapping for seed size and shape traits in soybean cultivars. Mol Breed 31:785–794
Osa M, Kato K, Mori M, Shindo C, Torada A, Miura H (2003) Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor Appl Genet 106:1491–1496
Sanguinetti CJ, Neto ED, Simpson AJ (1994) Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17:914–921
Schuurink RC, Van Beckum JMM, Heidekamp F (1992) Modulation of grain dormancy in barley by variation of plant growth conditions. Hereditas 117:137–143
Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, Yano M (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA 107:5792–5797
Takeuchi Y, Lin SY, Sasaki T, Yano M (2003) Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice. Theor Appl Genet 101:1174–1180
Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712
Ullrich SE, Clancy JA, del Blanco IA, Lee H, Jitkov VA, Han F, Kleinhofs A, Matsui K (2008) Genetic analysis of preharvest sprouting in a six-row barley cross. Mol Breed 21:249–259
Wan JM, Cao YJ, Wang CM, Ikehashi H (2005) Quantitative trait loci associated with seed dormancy in rice. Crop Sci 45:712–716
Wang Z, Chen Z, Cheng J, Lai Y, Wang J, Bao Y, Huang J, Zhang H (2012a) QTL analysis of Na+ and K+ concentrations in roots and shoots under different levels of NaCl stress in rice (Oryza sativa L.). PLoS One 7:e51202
Wang Z, Cheng J, Chen Z, Huang J, Bao Y, Wang J, Zhang H (2012b) Identification of QTLs with main, epistatic and QTL × environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor Appl Genet 125:807–815
Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22:1733–1748
Yang J, Hu CC, Ye XZ, Zhu J (2005) QTLNetwork 2.0. Available at http://ibi.zju.edu.cn/software/qtlnetwork. Institute of Bioinformatics, Zhejiang University, Hangzhou, China
Yasue T, Asai Y (1968) Studies on the dormancy and viviparous germination in rice seed. Res Bull Fac Agric Gifu Univ 26:1–12
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31271806; 31000748), the Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201203052), the Fundamental Research Funds for the Central Universities (Grant No. KYZ201202-9) and the 111 Project.
Conflict of interest
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding authors
Additional information
L. Wang and J. Cheng contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, L., Cheng, J., Lai, Y. et al. Identification of QTLs with additive, epistatic and QTL × development interaction effects for seed dormancy in rice. Planta 239, 411–420 (2014). https://doi.org/10.1007/s00425-013-1991-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1991-0