Abstract
Climate change will have major impacts on crop production: not just increasing drought and heat stress, but also increasing insect and disease loads and the chance of extreme weather events and further adverse conditions. Often, wild relatives show increased tolerances to biotic and abiotic stresses, due to reduced stringency of selection for yield and yield-related traits under optimum conditions. One possible strategy to improve resilience in our modern-day crop cultivars is to utilize wild relative germplasm in breeding, and attempt to introgress genetic factors contributing to greater environmental tolerances from these wild relatives into elite crop types. However, this approach can be difficult, as it relies on factors such as ease of hybridization and genetic distance between the source and target, crossover frequencies and distributions in the hybrid, and ability to select for desirable introgressions while minimizing linkage drag. In this review, we outline the possible effects that climate change may have on crop production, introduce the Brassica crop species and their wild relatives, and provide an index of useful traits that are known to be present in each of these species that may be exploitable through interspecific hybridization-based approaches. Subsequently, we outline how introgression breeding works, what factors affect the success of this approach, and how this approach can be optimized so as to increase the chance of recovering the desired introgression lines. Our review provides a working guide to the use of wild relatives and related crop germplasm to improve biotic and abiotic resistances in Brassica crop species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Climate change will result in a higher frequency of extreme weather events and increased pest and disease loads
Increasing concentrations of greenhouse gases resulting from industrial activity drive global warming via the greenhouse effect (IPCC 2014). Average temperatures are therefore rising globally, and are to date about 1 °C on average higher compared to pre-industrial levels (IPCC 2018), and about 1.5 °C higher over land (Shukla et al. 2019). Global temperatures will continue to rise a further 0.4–2.6 °C until 2050 depending on various climate protection policies (IPCC 2014). As a primary effect, rising temperatures increase the likelihood of heat waves (Shukla et al. 2019). Heat stress has negative impacts on plant growth due to its devastating influence on cell membranes and protein stability, and limits plant growth at all developmental stages, but particularly during flowering (Bita and Gerats 2013; Bailey-Serres et al. 2019). On top of direct effects, rising temperature can have two further adverse secondary effects on local climates: at warmer temperatures, the water holding capacity of the air increases about 7% per °C, which can lead to stronger single rain events and increase the likelihood of flooding (Trenberth 2011; Kodra et al. 2020). At the same time, rising transpiration can dry down soils more quickly and increase the likelihood of droughts (Trenberth 2011; Lu et al. 2019). Which outcome is more probable depends on season and geography. Central Europe, for example, can expect more rain in the winter season, but more drought in spring and early summer (Lu et al. 2019). Flooding leads to a loss of oxygen in the soil, which in turn leads to denitrification and ionic toxicity. Moreover, depending on how much of the plant is covered by water, flooding can also inhibit gas exchange and photosynthesis and therefore heavily impact plant metabolism (Sasidharan et al. 2018). Drought, on the other hand, leads to a loss of cell turgor, to which most crops react with closure of stomata (Iwaya-Inoue et al. 2018). This inhibits gas exchange and therefore leads to a loss in photosynthetic capacity (Chaves et al. 2009), with the production of reactive oxygen species as a negative side effect (Choudhury et al. 2017). Some farmers try to balance drought by increased irrigation when water resources are available, although this carries the risk of lowering ground water level and causing secondary salinification. The area of saline soils is also increasing, mostly due to unsuitable irrigation practices (Shukla et al. 2019), but also due to rising sea levels as a result of the ice shield melting and expansion of the oceans due to the warmer temperatures (Nerem et al. 2018; Cheng et al. 2020). Salinity negatively affects plant growth and survival, causing osmotic stress and ion toxicity (Chaves et al. 2009).
Finally, there are also tertiary effects of global warming. As climate zones start to shift (Shukla et al. 2019), insects and pests expand their climatic niche into higher latitudes and start spreading towards areas that were previously too cold for them (Suzuki et al. 2014). Moreover, increased abiotic stresses may weaken plant defense mechanisms against biotic stress (Suzuki et al. 2014).
The only putatively positive effect of rising industrial carbon dioxide levels is the fertilization effect via increased efficiency of the dark reaction of photosynthesis (Shukla et al. 2019). However, utilization of this effect depends on plant nitrogen and phosphorus availability (Sinclair et al. 2019) and is therefore mostly only expected in high-input farming. Moreover, the effect is expected to rapidly saturate due to the limited availability of RubisCO (Sinclair et al. 2019), such that additional rises in carbon dioxide are not going to increase growth further.
To summarize, the conditions for plant production are worsening quickly, and the available farm land is decreasing at the same time. Meanwhile, the global population is still rising, and we need to produce more food from less land and worse conditions than ever before. Therefore, crops need to be bred to produce more yield—we need to increase breeding gains. The major prerequisite for breeding gains is, however, genetic variation. In some crops, recent bottlenecks in breeding history have dramatically decreased genetic diversity within the gene pool, with Brassica napus (rapeseed) being a particular concern (Snowdon and Luy 2012). In this review, we introduce how Brassica wild relatives and the close relationships between crop species can be exploited to widen genetic diversity and improve resistances to biotic and abiotic stresses in this important group of crops, and outline potential methodology and considerations to using this approach in applied breeding programs.
The use of wild relatives and related species for crop improvement in Brassica
The Brassicaceae, also referred to as the mustard family or the Cruciferae, are a family of flowering plants comprising 338 genera and 3709 species (Al-Shehbaz et al. 2006; Warwick et al. 2006). The Brassicaceae contains several species of research interest, including the model plant Arabidopsis thaliana (hereafter referred to as Arabidopsis), as well as crops such as Raphanus sativus (radish), Eruca sativa (rocket), Sinapis alba (mustard seed), and Brassica napus (rapeseed). Some species such as Aurinia saxatilis (basket-of-gold), Iberis sempervirens (candytuft), Matthiola incana (stocks), Erysimum cheiri (wallflowers) and Lunaria annua (honesty) from this family are cultivated as ornamentals. The Brassiceae tribe is one of the 49 tribes in the Brassicaceae family, and is a group containing a number of phylogenetic lineages originating from a single clade. The Brassiceae contains species of various ploidy levels, with chromosome numbers for 80% of the species in this tribe ranging from n = 6 to n = 75 (Warwick and Anderson 1993). The genus Brassica, in the Brassiceae, is made up of 37 species and is the most agronomically significant genus in the Brassicaceae tribe, and has undergone extensive domestication (Gomez-Campo 1980). This genera includes mainly herbaceous plants believed to have originated from the Mediterranean region, and modern adapted cultivars have a global distribution as cultivated vegetables and oilseed crop plants (Fahey 2003). Brassica crops are commonly consumed as leafy (pak choy, kale), stem (wasabi) and root (turnip, swede, rutabaga) type vegetables, spice crops (black or brown mustard), cooking oil (rapeseed) and feed for livestock. Next in agronomic significance from the mustard family are Raphanus and Sinapis, which are also useful as edible roots (radish) and condiments (white mustard seeds) respectively (Rakow 2004). Owing to their closeness as members of the same Brassicaceae family, Brassica species benefit from the numerous molecular genetics and genomic tools available to Arabidopsis (Snowdon 2007; Mason and Snowdon 2016). The close relationship between species of the Brassica genus combined with the ample wild relatives and minor crop species in the wider Brassicaceae tribe make it an interesting model for examining interspecific hybridization for crop improvement (Katche et al. 2019).
The Triangle of U, developed by Korean cytogeneticist Nagaharu U (U 1935), shows the evolutionary and chromosomal relationships between the A, B and C genomes of the diploid species B. rapa (AA, 2n = 20; turnip rape, turnip, Chinese cabbage, Pak choi), B. nigra (BB, 2n = 16; black mustard) and B. oleracea (CC, 2n = 18; cabbage, cauliflower, broccoli, kale, kohlrabi, Brussels sprouts), and their allotetraploids B. carinata (AABB, 2n = 34; Abyssinian or Ethiopian mustard), B. napus (AACC, 2n = 38; oilseed rape, spring rape, swede) and B. juncea (BBCC, 2n = 36; Indian or brown mustard) which were generated through spontaneous interspecific hybridization events between the diploid species. Brassica napus is a relatively young crop (< 10 000 years old) which originated from the spontaneous hybridization between turnip rape (Brassica rapa; AA, 2n = 20) and cabbage/kale (Brassica oleracea; CC, 2n = 18) (Chalhoub et al. 2014). Wild types of B. nigra have been found in parts of Europe, Asia and North Africa (Oduor et al. 2015). As reviewed by Rakow (2004), B. nigra (n = 8, B genome) was initially identified as a weed in cultivated fields in the Mediterranean region, and is commonly seen on road sides and fields near Tangiers, Morocco and under semi-cultivated conditions in Rhodes, Crete, Sicily, Turkey and Ethiopia (Vaughan 1977; Tsunoda 1980). Brassica rapa (n = 10, A genome) originates from the highlands near the Mediterranean sea from where it migrated northward into Scandinavia and westward into eastern Europe and Germany (Nishi 1980). According to various authors, Brassica oleracea (n = 9, C genome), (characterized with distinct phenotypes (Snogerup 1980)), is believed to be a seaside plant of northern European or Mediterranean origin. Wild B. oleracea varieties still exist on maritime cliffs and continue to grow along the coasts of northern Spain, western France, southern and southwestern Britain (Vaughan 1977; Fahey 2003). Brassica carinata has been cultivated in Ethiopia and neighbouring territories from ancient times, while many researchers agree that B. juncea is a plant of Asiatic origin, with Asia as a centre of major diversity (Chen et al. 2013).
Rapeseed, oilseed rape or canola (Canadian Oil Low Acid) is the third most important oilseed crop in the world. Oilseed rape generally refers to any member of the Brassica genus which is grown for edible oil (normally B. napus, B. rapa and B. juncea), while rapeseed technically refers just to B. napus. Rapeseed attained economic importance as a source of edible vegetable oil after intensive breeding programs that led to the production of lines with low erucic acid (< 2% in the oil), low glucosinolate content (< 30 mg/g in the meal) and increased yields. All these breeding efforts and intensive selection for agricultural purposes have led to the generation of elite varieties with low genetic diversity compared to the wider gene pools (Snowdon and Luy 2012). Brassica napus, via human-assisted migration, went from Europe (where it first originated) to other parts of the world because of its usefulness as a high yielding Brassica crop with high seed quality (Zou et al. 2010). Winter rapeseed first spread to Russia, then to Japan and later on to China, while spring rapeseed reached China via Canada (Wu et al. 2019). Presently, almost 60% of the total global rapeseed production is from Canada, China and India (www.fao.org/faostat/November 2018), with the EU and Australia as other major rapeseed producers. In addition to serving as a good source of edible vegetable oil, rapeseed is also a valuable animal feed ingredient for ruminants and monogastric farm animals, used in producing industrial compounds like lubricants and surfactants and also as a raw material for biofuels in diesel cars and tractors, mostly in Germany and Europe (Allender and King 2010; Zou et al. 2010; Friedt et al. 2018). Qualities that make canola the preferred choice of oil by nutritionists and consumers around the world include its high content of poly-unsaturated linolenic acid (richness in omega-3, ca. 10%) and high content of oleic acid, ca. 60% (Iniguez-Luy and Federico 2011; Friedt et al. 2018). However, the balance of uses in the brassicas need to be maintained, as the value of the vegetable brassicas is outstripping B. napus globally, especially in light of rapeseed losses due to insects since the removal of important chemical controls by the European Union.
Useful traits identified in Brassica crops and wild allies
Each of the six major cultivated Brassica species contains unique, potentially useful agronomic traits that can be utilized to improve elite cultivars or to increase the gene pool within a species. While each species is often strongly associated with a particular phenotype, e.g. such that B. napus is widely known as a high yielding oilseed crop (76 MT produced in 2007, FAOSTATS) and B. oleracea as a highly variable vegetable type (Cheng et al. 2016), many traits present in individual species can be transferred between these closely related species for crop improvement. In the year 2009, a compendium of known traits in Brassica and wild relatives was published (Warwick et al. 2009). Since then, many other genotypes carrying relevant traits for agronomic improvement have been found in different Brassica accessions.
Disease resistance traits
Disease resistance has been broadly studied due to the major impact on crop production and yield. Resistance to a particular disease can be governed by a single gene (e.g. an “R-gene”) or by many genes with minor effects (quantitative resistance). Although many Brassica cultivars have been identified to carry particular disease resistances, pathogen evolution rapidly overcomes individual resistance sources or types under the high selection pressure of cropping production systems, such that the need for new resistance alleles is an ongoing process. Clubroot (CR) disease caused by many identified pathotypes of pathogen Plasmodiophora brassicae is prevalent around the world and greatly affects production in Brassica cultivars (Dixon 2009). Major resistance to CR has been found, for example, in Brassica rapa (Karling and Karling 1942; Piao et al. 2009; Zhang et al. 2015a). A large-scale screening for CR resistance against pathotype 3 carried out in a collection of 955 Brassica accessions (mostly B. rapa), revealed highly resistant accessions of B. rapa (17), B. nigra (4), and B. oleracea (2) (Peng et al. 2014). Another screening test of 22 CR isolates against 386 Brassica accessions (between 63–65 accessions of each species B. rapa, B. nigra, B. oleracea, B. napus, B. juncea, and B. carinata) revealed that most resistance sources were present in B. nigra, with some in B. oleracea, B. rapa and B. napus (but none identified in B. juncea or B. carinata) (Fredua‐Agyeman et al. 2019). Resistance to CR and downy mildew (Peronospora parasitica subsp. brassicae) was also tested in 52 accessions of B. oleracea and revealed frequent resistance to powdery mildew, but only a few lines possessed CR resistance (Carlsson et al. 2004). Further studies have also found field-based resistance to downy mildew in several B. oleracea lines (Monot and Silué 2009).
Sclerotinia stem rot (SSR), caused by Sclerotinia sclerotiorum, is a fungal disease that can cause considerable yield losses, with up to 70% infection incidence in winter oilseed rape when the conditions are suitable (Koch et al. 2007). Resistance for this disease has been identified in B. oleracea (Mei et al. 2011, 2013) and B. napus (Taylor et al. 2015), contrary to the high susceptibility found in B. juncea (Li et al. 2009). Recently, Sclerotinia resistance governed by several loci found in a wild C-genome species (B. incana) was introduced into B. napus via an interspecific hexaploidy hybrid bridge method (Mei et al. 2015, 2020). Through pyramiding three major QTLs, the BC1F8 line gained approximately 35% resistance when compared to the B. napus parent (Mei et al. 2020). Another strong source of resistance to SSR was found in Brassica fruticulosa (Rana et al. 2017). Subsequently, this resistance was transferred into a susceptible B. juncea genotype, producing introgressed lines with increased resistance, with a reduced lesion size of up to 69%. From the introgressed material it was also possible to select euploid and high pollen fertility lines, making it an excellent source to be utilized in future breeding programs (Rana et al. 2017).
Blackleg or phoma stem canker (caused by Leptosphaeria maculans) mainly affects rapeseed grown in Canada, Europe and Australia (West et al. 2001). One of the ways to control this disease is by sowing resistant cultivars; hence the need to find new genetic resources is always an ongoing process. Resistance to blackleg was found in lines of B. napus (Delourme et al. 2006; Rimmer 2006; Light et al. 2011) and B. rapa subsp. sylvestris (Yu et al. 2005, 2008). To date, no resistance R gene against blackleg has been observed in the C Brassica genome, although some possible in silico candidates have recently been proposed (Ferdous et al. 2020). Other kinds of resistances that involve more than just one gene are known as quantitative disease resistances. This resistance type is associated to particular genomic region/s or quantitative trait loci (QTL) that contribute to a partial level of disease resistance, usually more complex to identify due to its nature, but in the long term, harder for the pathogen to overcome (Pilet-Nayel et al. 2017). Several blackleg resistance QTL have been identified in spring-type Brassica napus (Larkan et al. 2016) and in diversity sets of Brassica napus (Jestin et al. 2011; Rahman et al. 2016; Raman et al. 2016).
Brassica oleracea is the major host for black rot (Xanthomonas campestris pv. campestris) (Vicente et al. 2001). This disease can cause severe damage, affecting up to 50% of the crop (Singh et al. 2011). Several resistant lines have been found in B. oleracea (Lema et al. 2012; Saha et al. 2016; Ribeiro da Silva et al. 2020) and B. rapa (Lema et al. 2015). In search of resistance in other subgenomes than the C, a single gene resistance locus was identified in B. carinata, located on linkage group B7 (Sharma et al. 2016). Later on, this resistance was introgressed into B. oleracea using embryo rescue (Sharma et al. 2017).
Resistance to white rust (WR) caused by the pathogen Albugo candida has been found in B. juncea, B. napus, B. rapa and B. carinata varieties (Panjabi-Massand et al. 2010; Awasthi et al. 2012). Quite recently, a B. juncea Chinese vegetable type mustard called Tumida was found to be resistant to WR, for which a responsible locus was located on linkage group A06 (Bhayana et al. 2020). Different Brassica genotypes and allies from diverse origins were tested against Pseudocercosporella capsellae (white leaf spot disease) in field and/or controlled conditions, and genotypes from B. carinata, B. juncea, B. napus, B. oleracea and B. fruticulosa shown to be highly resistant (Gunasinghe et al. 2014, 2017). By comparing resistant and susceptible lines derived from the three allotetraploid Brassica types, Gunasinghe et al. (2016) identified a resistant B. carinata line possessing stomata prone to closure to inhibit pathogen penetration. Also, a higher stomata density was observed in the susceptible lines.
Insect resistance traits
Insects also are a big problem in Brassica crops and major yield losses and aesthetic damage can occur under their attack. The major pests attacking Brassica belong to the order of Lepidoptera, Hymenoptera, Diptera, Homoptera and Coleoptera (reviewed in (Ahuja et al. 2011)), many of them with the ability to move and migrate to infest their hosts. A very common oilseed rape pest is the pollen beetle (Brassicogethes aeneus, previously known as Meligethes aenus), that can cause more than 80% yield losses (Hansen 2004). Unfortunately, to date, no natural resistance has been found and the only way to protect the plants is through insecticide application or other integrated pest management strategies. Due to this, new resistant insects have emerged (Spitzer et al. 2020) and novel strategies are required to control the pest (reviewed in (Hervé and Cortesero 2016)). For other pests, such as Diamondback moth (Plutella xylostella), that can cause severe economic damage (Zalucki et al. 2012; Li et al. 2016), resistance has been observed in a single line of B. oleracea spp. capitata (Kim et al. 2013).
In 2015, 432 different accessions of B. oleracea and allies were tested against cabbage whitefly (Aleyrodes proletella L.), out of which 48 showed a high degree of resistance (Pelgrom et al. 2015). In this study, the wild relatives B. incana, B. montana and B. villosa were shown to be very unappealing to the pest under early growth and development conditions. One possible explanation for the observed resistance in B. incana is the presence of trichomes (absent in the susceptible genotype).
The pest known as cabbage seedpod weevil (CSW; Ceutorhynchus obstrictus) severely affects oilseed rape, especially during the early flowering period (reviewed in (Dosdall 2009)). Resistance for this pest has been found in lines produced by the cross of Sinapis alba (resistant parent) and B. napus (susceptible parent) (Tansey et al. 2010; Lee et al. 2014). Resistance to another weevil pest, Ceutorhynchus napi, also known as rape stem weevil, was found in resynthesized B. napus lines (Schaefer-Koesterke et al. 2017). The resistance observed might be due to antixenosis (non-preference) given the extended size of the stem and also the lack of specific glucosinolate compounds (Schaefer-Koesterke et al. 2017).
Fully developed cabbage root fly (Delia radicum L.) infests its host by laying eggs on the ground, close to the plant, where the larva can live by feeding from the roots, therefore affecting plant development and eventually damaging yield loss (Hopkins et al. 1999). In a panel composed of diverse Brassica species, the antibiosis resistance (adverse effects on the pest) of these plants against cabbage root fly was studied (Shuhang et al. 2016). Here they found high levels of antibiosis in B. spinenscens and B. fruticulosa under greenhouse conditions, given by the observed fewer eclosed flies per egg and reduced fly dry weight (Shuhang et al. 2016). Other potential resistance candidates more readily crossable with Brassica crops are accessions found in B. montana, B. macrocarpa, B. villosa, B. hilarionis (Shuhang et al. 2016) and B. rapa (Santolamazza-Carbone et al. 2017).
In a 2-year case study, Eruca sativa cv. T 27 followed by B. carinata cv. DLSC 2 were the least infested by aphids (Lipaphis erysimi) under normal conditions when compared to B. juncea, B. rapa and a hybrid B. napus (Kumar and Sangha 2017). Between the species studied, there were different chemical profiles present in the inflorescence that can explain over 94% of the amount of aphids present (Kumar and Sangha 2017). Screening for resistance to the moth Mamestra brassicae was carried out in 21 cabbage (B. oleracea var. capitata) varieties (Cartea et al. 2010). The two more resistant varieties had the compact head characteristic, a morphological trait that can also be involved in insect resistance (Carmona et al. 2011). Some of the insect pests affecting Brassica plants can work as carriers of other diseases like viruses. There are several viral infections described as affecting Brassica crops, especially cabbage types, including for example cauliflower mosaic virus (CaMV), turnip yellow mosaic virus (TyMV) and turnip mosaic virus (TuMV) (Raybould et al. 1999). A combination of TuMV and CaMV infection can affect up to 25% of the yield in B. oleacea var. capitata, mostly due to TuMV, as no significant effect was observed when CaMV was inoculated alone (Spence et al. 2007), and in current times most research has focused on identifying resistance for TuMV. Turnip mosaic virus (TuMV) infection in crucifer plants was initially described in 1921 (Schultz 1921), where the characteristic spotted pattern of a “mosaic like virus” was observed in Brassica rapa. This disease is mainly transmitted by aphids and non-exclusively infects Brassica genotypes (Walsh and Jenner 2002; Shattuck 2010), causing a reduction in fitness, reproduction and quality of the plant (Maskell et al. 1999). The utilization of insecticides against aphids to control the spread of TuMV is not very efficient; consequently the identification and utilization of naturally resistant Brassica varieties becomes the preferred option to control the disease in an environmentally friendly way (Walsh et al. 1999).
In one study, B. juncea, B. oleracea, B. rapa, C. sativa and R. sativus lines were tested against TuMV virus pathotype 8 (Nyalugwe et al. 2015): different B. oleracea and R. sativus lines showed consistently extreme resistance to the virus. The rest of the lines showed different responses to the infection although there was potential for resistance in each of the species tested (Nyalugwe et al. 2015). Also in this study, a dominant gene conferring systemic resistance in B. juncea was identified (TuMV RESISTANCE IN BRASSICA JUNCEA 01) (Nyalugwe et al. 2015, 2016). Extreme resistance to the TuMV pathotype 8 has been also observed in 18 B. napus and 14 B. carinata lines from different origins (Nyalugwe et al. 2014). A resistance to TuMV virus found in Raphanus sativus was identified and successfully transmitted via somatic fusion with B. oleracea var. capitata, B. oleracea var. botrytis, B. oleracea var. capitata, to 61, 83.6 and 33.2% of the hybrids produced, respectively (Scholze et al. 2010).
Abiotic stress tolerances
Abiotic stress tolerances also vary across the Brassica species. Salt tolerance has been shown to be greater in the allopolyploids B. juncea, B. napus and B. carinata than in their diploid parents (Ashraf et al. 2001). Similar effects were observed when comparing salinity tolerance between various Brassica genotypes and ploidies (Kumar et al. 2009). In a different study, where tetraploid turnips (B. rapa) were compared to diploid progenitors, it was also shown that this increase in ploidy positively affects salinity tolerance (Meng et al. 2011). A wide diversity set of B. napus accessions (85 inbred lines) were tested for salt tolerance under hydroponic conditions (Yong et al. 2015). The results showed significant variation in shoot fresh weight and dry weight between the different accessions and, at the same time, there was no correlation between sodium ion accumulation in leaves and the salt tolerance index.
Screening of nine different B. juncea genotypes resulted in the discovery of one tolerant genotype (Varuna) among them (Hayat et al. 2011). In B. juncea, several other tolerances have been observed, such as heat stress (Wilson et al. 2014) and cadmium tolerance (Gill et al. 2011; Irfan et al. 2014). Nevertheless, many of the results obtained for tolerance to heavy metals depend on the methods utilized to screen the tolerance (Hernández-Allica et al. 2008) and also not all of them are easily comparable due to these differences (reviewed in (Mourato et al. 2015)).
Polluted soil, water or air can be of great danger to human health. Fortunately, we can use plants to remove those contaminants, a term known as phytoremediation (reviewed in Salt et al. 1998). An excellent example of this is B. juncea var. foliosa, which has the potential to be used in phytoremediation in thorium (Th) contaminated soils due to its ability to tolerate this metal (Zhou et al. 2016). Under low concentrations of Th, B. juncea var. foliosa grew better, but under high concentrations plant metabolism and growth rates were affected. Some of the Brassica vegetable types, like Brassica rapa subsp. pekinensis (Chinese cabbage), also have the capacity to accumulate high amounts of heavy metals without any obvious symptoms, presenting a potential risk for human food contamination (Xiong and Wang 2005).
Lack of water during flowering can heavily impact the final yield production of plants. Thankfully, we can use the available germplasm of a species to investigate how well they are able to cope, and even recover if they were submitted to water stress. Phenotyping for drought stress tolerance in B. napus under simulated normal and osmotic stress conditions in a hydroponic system combined with GWAS revealed 16 stress-tolerant accessions and 16 SNP loci associated with osmotic stress response (Zhang et al. 2015b). When comparing single genotypes of B. napus, B. rapa and B. juncea under simulated drought stress (using PEG-6000), it was found that B. juncea was more drought tolerant than the other two species (Alam et al. 2014). A study of drought tolerance in B. napus pre- and post-flowering conditions found 3 and 4 different accessions tolerant to drought, respectively (Zhu et al. 2011). A closer characterization of drought tolerance mechanisms in B. napus revealed that individual strategies vary strongly between accessions, but common drought tolerance genes might exist (Schiessl et al. 2020).
Other traits of agronomic interest
A number of other miscellaneous traits of agronomic importance are also present in various Brassica species. Cytoplasmic male sterility (CMS) is a widely utilized system to produce F1 hybrids in Brassica crops taking advantage of the high hybrid vigour observed in seed yield (Yamagishi and Bhat 2014). Several systems have been found in species like B. juncea (hau CMS, (Wan et al. 2008)), B. napus (nap and pol CMS, (Brown 1999)), B. rapa (YSMS-6 (Bhajan 2000); eru CMS (Peng et al. 2015)), Raphanus sativus (ogu CMS (Ogura 1968)), B. oleracea (Zhiyuan et al. 1995; Fang et al. 1997) and a system produced by the cross between B. napus and B. carinata (NCa (Wei et al. 2009)).
Leaves are very important organs, where processes like photosynthesis, respiration and transpiration take place, and also are the initial barrier against environmental conditions. Leaf composition can also act as a barrier against herbivore attack (Žnidarčič et al. 2008; Bohinc et al. 2014). In B. juncea leaf morphology was studied in 10 wild accessions (Huangfu et al. 2009): the different populations varied in leaf thickness, wax content, and leaf surface, among other morphological traits. Interestingly, some of the phenotypes analyzed also correlated with herbicide (glyphosate) resistance in the populations, especially leaf thickness, with an R2 of 0.72.
Pod shattering, from an evolutionary point of view, is a great mechanism for seed dispersal. Unfortunately, from an economical point of view, in Brassica oilseed crops it can cause great seed losses during harvesting, which under normal conditions can reach up to 2–5%, and when the conditions are less than optimal values over 20% or up to 50% can be obtained (Price et al. 1996). Pod shatter resistance is present naturally in B. carinata, B. juncea and B. rapa genotypes (Raman et al. 2014). On the other hand, the variation present in B. napus for pod shattering is more limited. For example, in a study where 229 B. napus accessions were investigated for silique shattering resistance, just two varieties were fully resistant (Wen 2008).
Novel traits may also be utilized to produce niche Brassica types for different purposes. For instance, Brassica species are characterized by the ubiquitous presence of glucosinolates, although the amount and composition of these compounds varies depending on the tissue or cultivar analyzed (Verkerk et al. 2010; Sun et al. 2019). Glucosinolates are secondary metabolites that have been associated in plants with insect resistance (Evivie et al. 2019), fungal resistance (Bednarek et al. 2009; Buxdorf et al. 2013), signalling molecules in the auxin pathway (Katz et al. 2015), its involvement in other biological processes like flowering time and stomatal closure (reviewed in (Barco and Clay 2019)) and even the possible contribution in preventing certain types of human cancer like lung, stomach and prostate when included in the diet (reviewed in (Traka and Mithen 2009)). Most commercial oilseed Brassica cultivars have been bred to contain low levels of glucosinolates, which is more desirable for edible oil. However, there is a niche for specific glucosinolate profiles that are desirable for other applications, for example in industrial oil production (Princen 1979).
Carotenoid content is another trait of interest that can potentially be manipulated and bred to produce edible plants with specific profiles, to fit human needs. In B. oleracea, diverse carotenoid composition was observed in a set of 30 different cultivars from various origins (Mageney et al. 2016). In B. rapa spp. pekinensis, a hybrid produced by the cross of two incompatible cultivars produces a hybrid with orange inner leaves (Yangjun et al. 2005). Interestingly, also in B. rapa cultivars, the production of other pigments (anthocyanin) has been associated with cold and freezing resistance (Ahmed et al. 2015).
Hybridization for crop improvement in Brassica
Genetic diversity within and between species is a prerequisite for breeding and crop improvement. In order to improve yields, increase disease resistance and refine oil qualities to cater to various nutritional and industrial purposes, it is imperative to introduce new sources of genetic diversity into existing elite cultivars (Allender and King 2010). In the Brassicaceae, new variation can be generated by hybridization involving adapted cultivars, wild types and landraces or exotic germplasm such as different species (Friedt et al. 2018).
Interspecific hybridization is useful in the introgression of desirable traits from one species to another and there are different approaches for transferring traits through interspecific hybridization (Prakash et al. 2009; Mason and Chèvre 2016). The success of crosses between any two parents can be determined by observing their pollen germination, pollen tube growth, embryo development, and seed set (Bhat and Sarla 2004). Hybrid incompatibilities occur when hybrids are sterile, less fit or even non-viable compared to their progenitors: this serves as a reproductive isolation barrier which can lead to speciation (Coyne and Orr 2004). Within the Brassica genus, incompatibilities may occur between different species, cultivars or species with different ploidy levels (Nishiyama et al. 1991; FitzJohn et al. 2007). To date, a number of genes with diverse functions, including those involved in oxidative respiration, nuclear trafficking, DNA-binding, and plant defence have been linked to hybrid incompatibilities (Johnson 2010; Rieseberg and Blackman 2010), but the underlying genetic and molecular mechanisms are not yet fully understood (Vaid and Laitinen 2019).
Other mechanisms that prompt hybrid incompatibility include conflicts resulting from the unequal parental contribution to the formation of hybrid or developing seed (Carputo et al. 2003; Johnson 2010; Köhler et al. 2010). This is often seen in the different phenotypes or success rate obtained when reciprocal crosses are made. In crosses between Brassica species, the choice of maternal species has a big effect on the success of the cross (FitzJohn et al. 2007; Chen et al. 2011). Another well-known example is the cytoplasmic male sterility (CMS) which is a maternally inherited trait characterized by the inability of a plant to produce functional pollen (Eckardt 2006). Hybrid chlorophyll deficiency causes white-coloured cotyledons and this has been identified to occur as a result of incompatibility between the plastid genome and the nuclear genome (Ureshino et al. 1999; Okamoto and Ureshino 2015).
Hybrid necrosis or death of young seedlings is another form of post-zygotic incompatibility which is associated with complexities in gene interaction (Potts and Dungey 2004; Okamoto and Ureshino 2015). In Arabidopsis for instance, it has been revealed that conflict between two gene variants or loci (DANGEROUS MIX 1 (DM1) and DANGEROUS MIX 2 (DM2)) may trigger defence reactions which can be detected phenotypically in hybrids as necrotic lesions on leaves and a decline in growth and fertility (Bomblies and Weigel 2007; Chae et al. 2014). ACCELERATED CELL DEATH 6 (ACD6) is another gene that causes hybrid necrosis when its allele variants interact leading to the activation of pathogen-recognition receptors and trigger autoimmune response to pathogens in first generation hybrids of A. thaliana (Todesco et al. 2014; Tateda et al. 2015; Świadek et al. 2017).
Even after successful pollen germination and fertilization, the abnormal growth of the endosperm can interfere with normal seed development (Haig and Westoby 1991; Lafon-Placette and Köhler 2016). Similarly, in Brassica species, interspecific hybridization does not always lead to the production of mature seeds, as a result of irregularities in endosperm development (Nishiyama et al. 1991). Failure of endosperm development in hybrids may occur as a result of unbalanced parental genome dosages or genomic imprinting (Köhler et al. 2010).
Transferring useful traits from wild relatives to crop species: how does it work?
Although a major QTL PrBn (Pairing regulator in B. napus) and other minor QTL have been observed to affect non-homologous chromosome pairing frequencies in Brassica napus allohaploids (Jenczewski et al. 2003; Liu et al. 2006), Brassica species generally have weak, quantitative regulation of meiosis, which readily permits hybridization and introgressions to transfer useful traits between genomes (Fig. 1).
(adapted from Mizushima 1980): Genome interrelationships in Brassica and allied genera. Numbers in brackets represent the number of autosyndetic bivalents observed in haploids, while numbers on lines indicate the maximum number of bivalents observed in interspecific hybrids between the two species (necessary for transferring traits between genomes)
The first step in transferring useful traits from wild relatives to crops is to identify which wild relative germplasm carries the trait of interest, and preferably also the genetic basis for this trait. Ideally, the target germplasm will be within the same species, and the trait will be carried by a single major gene locus. Unfortunately, this situation is rarely found. Firstly, many species are relatively inbred, lacking the genetic and trait diversity necessary for further specific improvements. In the Brassica genus, this is particularly true in major crop species B. napus (rapeseed), for which no “wild” forms exist (Dixon 2007), and in which (for example) little to no resistance to insect predation is thought to exist (Hervé 2018). Hence, it is often necessary to look outside this so-called primary germplasm pool for traits. Secondly, although some traits are often carried by major genes, such as resistance to blackleg/Phoma disease (Rimmer 2006; Leflon et al. 2007) or resistance to clubroot (Manzanares-Dauleux et al. 2000), most traits, including drought tolerance (Fletcher et al. 2015, 2016; Zhu et al. 2016), flowering time (Schiessl et al. 2014, 2015), and of course yield (Zhou et al. 2014; Luo et al. 2017), tend to be the product of multiple genes, genetic factors or gene networks.
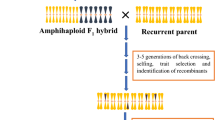
The reason that it is better to have traits which are (a) present in closely-related species and (b) controlled by a single locus is because of the mechanisms by which we transfer traits from the wild to crop germplasm. The physical transfer of genetic material between two germplasm groups usually needs to occur via one or more crossovers between chromosomes in the hybrid which has been produced between them, which (usually) has 50% genetic material from each parent (Fig. 2).
While it is relatively easy to make hybrids within a species by hand-emasculation and pollination, this becomes much more difficult with increasing genetic distance between the wild relative and the crop (FitzJohn et al. 2007). As well, the subsequent chance of recovering recombination events is greatly reduced if there is little relationship between the two sets of chromosomes present in the hybrid, such that they rarely pair and recombine with each other (Mason and Chèvre 2016)(Fig. 1). If multiple genetic loci need to be transferred, even more crossovers need to form, and this further reduces the chance of recovering the desirable trait in segregating hybrid progeny (Mason and Chèvre 2016). Depending on the genomic location of the locus of interest, it may not even be possible to produce recombinants through conventional means, as crossovers are not evenly distributed across chromosomes (reviewed by (Choi and Henderson 2015)), and are actively suppressed in others, such as centromeres (Talbert and Henikoff 2010). Hence, some genomic regions are very unlikely to recombine during meiosis: when considering two target regions in a hybrid, the probability of a natural crossover forming between these two may be so low as to be effectively non-existent. Also, every transfer has the potential to introgress large blocks of undesirable genetic variation as well as the desirable genetic variation conferring the trait of interest (linkage drag), as normally, a large chromosomal segment will be introgressed from a single crossover. In the case of intraspecific crosses or crosses between species with very high genomic similarity this is not such a big problem: subsequent recombination events may occur through backcrossing to the crop parent, and thus reduce the size of the introgression block (Fig. 2). This eliminates undesirable genetic variation while retaining the locus of interest. However, further recombination events cannot be guaranteed in the case of wide crosses, which may mean that the resulting introgression region is large and carries a high number of undesirable genetic variants. This problem was classically encountered in Brassica breeding with the production of the restorer lines for the “Ogura” CMS system developed from radish wide hybrids (Pellan-Delourme and Renard 1988), and for which gamma ray induction of chromosome breakage was required to reduce introgression size (Primard-Brisset et al. 2005).
Improving our chances of recovering introgressions of useful traits from relatives into crops to build climate resilience
So, what can we do to facilitate transfer of genetic loci and traits of interest from wild relatives into crops to build climate resilience? Good experimental planning and prior knowledge is key to improving success rates. Although in some cases very little is known about (a) genetic control of the target trait or phenotype in question, (b) ease of hybrid production, (c) frequency and distribution of crossovers in the interspecific hybrid or (d) chance of recovering successful introgressions, most of the time at least some of this information should already be known, and can be used to predict the amount of time and effort likely required to achieve this goal. Recent developments in genomics and bioinformatics techniques are predicted to help a lot in this respect (for review see (Zhang and Batley 2020)). However, there are also a number of specific methods or considerations that can be used to facilitate this process.
Hybrid generation is not always successful, especially across different ploidy levels. However, a number of crossing approaches can be used to facilitate trait transfer through interspecific hybridization in Brassica species and their relatives (Prakash et al. 2009; Mason and Chèvre 2016). Hybridization is generally more successful between species which share a genome, e.g. between a tetraploid and a diploid progenitor species, or between two tetraploids which share a progenitor ((Prakash et al. 2009); reviewed in Mason and Chèvre 2016). Hybridization between diploids and tetraploids that do not share a genome is also possible, and the resulting tri-genomic hybrids can be used as a bridge to introgress genetic diversity between species in further hybridization events, or can be induced by colchicine doubling to generate allohexaploids (Chen et al. 2011). While interspecific hybridization is very useful for hybrid speciation and crop improvement in the Brassica genus, hybridizations can also be made between genera. Hybridization involving the Brassica crop species is often successful using only hand-pollination methods (FitzJohn et al. 2007). However, hybridization can be facilitated by tissue culture techniques which “rescue” fertilized ovules or embryos before these are aborted by the maternal parent (reviewed by Sharma et al. 1996). For wider crosses somatic fusion may also be possible, where somatic cells (usually protoplasts) are directly induced to combine in tissue culture (reviewed by (Navrátilová 2004)), although this method frequently results in aneuploidy (loss or gain of individual chromosomes from a set) (Gaebelein and Mason 2018). A common example is the protoplast fusion of rapeseed and radish. This method was employed in generating the Ogura cytoplasmic male sterile B. napus system and was very beneficial in attaining double low restorer lines with a decrease in erucic acid and glucosinolate content (Pelletier et al. 1983; Primard-Brisset et al. 2005). Better success may also be achieved in some cases by chromosome doubling the parent species before hybridization is attempted (Frandsen 1947; Heyn 1977; Akbar 1990); this can also be achieved by various chemical treatments and methods (reviewed by (Dhooghe et al. 2011)). Other approaches used in tackling these incompatibilities include hot water treatments against pre-fertilization barriers (Prabha et al. 1982), early pollination of stigmas or stump pollination, in vitro pollination (reviewed in Katche et al. 2019), and artificially supplied nutrients and hormones against post-fertilization barriers (Sharma et al. 1996; Abel et al. 2005; Prakash et al. 2009).
Once a hybrid is produced, every meiosis in this hybrid (every pollen or ovule produced) has the potential to produce recombinant chromosomes (introgressions) between the source and target genomes. Meiotic recombination is an important aspect of breeding, as it ensures plant fertility and the generation of diversity through shuffling of genetic information. However, CO localization is uneven across the genome, with 80% of all COs occurring in about 25% of genomic regions in most plants (usually at the distal euchromatic regions) (Darrier et al. 2017). Obtaining the meiotic recombination required for crop improvement is also challenging in plants with low CO frequencies. Knowledge of how often recombination events occur in different types of hybrid is invaluable in knowing approximately how many progeny may need to be obtained in order to recover the desired introgression. Even better, knowledge about genome-wide distribution of recombination rates in hybrids of different types would allow predictions of success in introgression of particular genetic loci before experiments even start. However, COs can potentially be increased through several mechanisms, including the knockout of anti-crossover regulators such as FANCM, RECQ4 and FIGL1, or combining the knockout of anti-CO regulators with an increase in the dosage of ZMM protein HEI10, and through mutagenesis approaches (Blary and Jenczewski 2019). In Brassica allotriploid AAC hybrids, there is an increase in CO rates between the homologous A chromosomes (Leflon et al. 2010); better understanding and characterization of this effect may be helpful in applying this crossover boost to other hybrid types. In future, it may also be possible to manipulate the regulation of chromosome pairing between genomes in order to boost the frequency of crossovers and change their genomic locations (reviewed by Blary and Jenczewski 2019).
To facilitate introgressions in the absence of other information about recombination frequencies or crossover distributions, target genes should ideally be close to chromosome telomeres and in chromosomal regions with high homoeology (or homology if possible) between the two sets of chromosomes in the hybrid. In B. juncea × B. carinata BBAC hybrids, A-C pairing is extraordinarily frequent, with an average of 7 A-C allosyndetic pairs per meiosis (Mason et al. 2010), almost always between primary homoeologous regions (Mason et al. 2014). In the same hybrids, autosyndetic recombination events (A-A and C–C in the haploid genomes) occur at a rate of approximately one event per 2 meioses (Mason et al. 2010), but likely only between the largest blocks resulting from the ancestral genome triplication, involving up to a chromosome arm (Mason et al. 2014). In hybrids resulting from the cross B. napus × B. nigra, B-A/C allosyndesis is observed in 1/3 meioses in ABC triploid hybrids, and 1/6 meioses in AABBCC allohexaploid hybrids (Gaebelein et al. 2019), although which genomic regions are recombining is not known. Although hypothetically any genomic similarity can trigger recombination, crossovers have strong “preferences”, and will form between whatever chromosomes are present on order of sequence similarity first (Grandont et al. 2014). In the absence of homologous pairing partners, recombination will occur most frequently between the most closely-related (or possibly largest) homoeologous regions (Nicolas et al. 2007, 2009; Mason et al. 2014). Regions of primary and secondary (resulting from ancestral triplication) homoeology have been well-defined for quite some time for the A and C genomes (Parkin et al. 2003; Schranz et al. 2006; Cheng et al. 2013), and to a lesser extent the B genome (Lagercrantz and Lydiate 1996). Now, with the availability of reference genome sequences, these relationships have been even better elucidated (The Brassica rapa Genome Sequencing Project Consortium 2011; Chalhoub et al. 2014; Parkin et al. 2014; Yang et al. 2016; Wang et al. 2019), and subsequently can be used to predict the probable locations of genomic introgressions, even though we are still lacking a lot of information about where crossovers are most likely to form.
Conventionally, we can boost our chances of transferring desirable loci by selecting good targets: single, major gene effects, present in close relatives to our crop of interest. However, several agronomic traits of interest, including yield, plant height and flowering time, are controlled by many genes and heavily influenced by the environment, and thus present a greater challenge. The genetic control or mode of inheritance of a desired trait can be examined through marker analysis or by observing how traits segregate in progenies. Owing to the recent advancement in genotyping, gene editing and marker technologies, the characterization and introgression of our gene of interest, genomic regions (complex traits) or even pyramiding of multiple QTLs can readily be done. In a number of studies, introgression was successfully achieved via a combination of different hybridization schemes. In general, creation of a suitable mapping population to elucidate the genetic control of the trait, followed by association of phenotypes with genotypes to identify genomic regions of interest and subsequent development of marker assisted selection (MAS), is an excellent strategy to facilitate production of introgression lines. For instance, to introgress Sclerotinia resistance into rapeseed, Mei et al. (2020) transferred multiple resistant loci from wild B. oleracea through backcrossing, selfing and MAS. A different approach was used by Mei et al. (2015) to introgress Sclerotinia resistance from B. incana (a wild relative of Brassica oleracea) into B. napus bridged by a hexaploidy step.
Tracking of introgressions can be sped up by marker-assisted selection, or possibly even by genomic selection in the case of multi-locus traits being transferred between close relatives. Marker-assisted selection has been effectively used to track B-genome introgressions related to Sclerotinia disease resistance in Brassica napus (Navabi et al. 2010), to produce higher-quality restorer lines carrying the Rfo restorer gene for the Ogura CMS system in B. juncea by reducing the size of the radish introgression (Tian et al. 2014), and to map and move clubroot resistance gene Rpb1 from B. rapa into B. napus (Chu et al. 2013). Genome-wide marker assisted-selection has also been used in several studies to recover subgenome-substitution or resynthesized lines. This approach has been used to produce “new-type” B. napus (AACC) with an A genome from B. rapa (Ar) and a C genome (Cc) from B. carinata (Xiao et al. 2010), through selection for Ar and Cc alleles over An and Cn, as well as to extract the B. napus A genome by eliminating the C genome (Pelé et al. 2017). Hence, genome-wide marker-assisted selection may be worth considering in the case of complex traits which are being moved between species which share a subgenome (e.g. species within the Triangle of U).
Conclusions
In this review, we introduce potential impacts of climate change on crop production and the Brassicaceae crops, provide a reference for useful traits present in each of the Brassica “Triangle of U” species and then offer concrete advice for structuring and optimizing introgression breeding programs. Success in transferring agronomically relevant traits between species depends on factors such as similarity between the source (e.g. wild relative) and target (e.g. crop) genomes, the ease of hybrid production, the frequency and distribution of crossovers in the interspecific hybrid meiosis, and subsequently the ease of recovery of introgression lines. Regardless of the considerable difficulties involved in the use of wild relatives for crop improvement, this method offers a great deal of as-yet unexplored potential for the improvement of Brassica crops, and in improving crop resilience and resistances in the face of global climate change.
References
Abel S, Möllers C, Becker H (2005) Development of synthetic Brassica napus lines for the analysis of “fixed heterosis” in allopolyploid plants. Euphytica 146:157–163
Ahmed NU, Park J-I, Jung H-J et al (2015) Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct Integr Genom 15:383–394. https://doi.org/10.1007/s10142-014-0427-7
Ahuja I, Rohloff J, Bones AM (2011) Defence mechanisms of brassicaceae: implications for plant-insect interactions and potential for integrated pest management. Sustainable Agriculture, vol 2. Springer, pp 623–670
Akbar MA (1990) Resynthesis of Brassica napus aiming for improved earliness and carried out by different approaches. Hereditas 111:239–246. https://doi.org/10.1111/j.1601-5223.1990.tb00402.x
Al-Shehbaz IA, Beilstein MA, Kellogg EA (2006) Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst Evol 259:89–120. https://doi.org/10.1007/s00606-006-0415-z
Alam MM, Nahar K, Hasanuzzaman M, Fujita M (2014) Alleviation of osmotic stress in Brassica napus, B. campestris, and B. juncea by ascorbic acid application. Biol Plant 58:697–708. https://doi.org/10.1007/s10535-014-0447-0
Allender CJ, King GJ (2010) Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol 10:54. https://doi.org/10.1186/1471-2229-10-54
Ashraf M, Nazir N, McNeilly T (2001) Comparative salt tolerance of amphidiploid and diploid Brassica species. Plant Sci 160:683–689. https://doi.org/10.1016/S0168-9452(00)00449-0
Awasthi RS, Nashaat NI, Kolte SB et al (2012) Screening of putative resistant sources against Indian and exotic isolates of Albugo candida inciting white rust in rapeseed-mustard. J Oilseed Brassica 3:27–37
Bailey-Serres J, Parker JE, Ainsworth EA et al (2019) Genetic strategies for improving crop yields. Nature 7781:109–118. https://doi.org/10.1038/s41586-019-1679-0
Barco B, Clay NK (2019) Evolution of glucosinolate diversity via whole-genome duplications, gene rearrangements, and substrate promiscuity. Annu Rev Plant Biol 70:585–604. https://doi.org/10.1146/annurev-arplant-050718-100152
Bednarek P, Piślewska-Bednarek M, Svatoš A et al (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323:101–106. https://doi.org/10.1126/science.1163732
Bhajan R (2000) Development of YSMS-6, a genetic male sterile line of yellow sarson (Brassica campestris var yellow sarson Prain). Crucif Newsl Eucarpia 22:29–30
Bhat S, Sarla N (2004) Identification and overcoming barriers between Brassica rapa L. em. Metzg. and B. nigra (L.) Koch crosses for the resynthesis of B. juncea (L.) Czern. Genet Resour Crop Evol 51:455–469. https://doi.org/10.1023/B:GRES.0000024154.19867.cd
Bhayana L, Paritosh K, Arora H et al (2020) A mapped locus on LG A6 of Brassica juncea line tumida conferring resistance to white rust contains a CNL Type R Gene. Front Plant Sci 10:1690. https://doi.org/10.3389/fpls.2019.01690
Bita CE, Gerats T (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4:273. https://doi.org/10.3389/fpls.2013.00273
Blary A, Jenczewski E (2019) Manipulation of crossover frequency and distribution for plant breeding. Theor Appl Genet 132:575–592. https://doi.org/10.1007/s00122-018-3240-1
Bohinc T, Markovič D, Trdan S (2014) Leaf epicuticular wax as a factor of antixenotic resistance of cabbage to cabbage flea beetles and cabbage stink bugs attack. Acta Agric Scand Sect B Soil Plant Sci 64:493–500. https://doi.org/10.1080/09064710.2014.926978
Bomblies K, Weigel D (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8:382–3893. https://doi.org/10.1038/nrg2082
Brown GG (1999) Unique aspects of cytoplasmic male sterility and fertility restoration in Brassica napus. J Hered 90:351–356. https://doi.org/10.1093/jhered/90.3.351
Buxdorf K, Yaffe H, Barda O, Levy M (2013) The effects of glucosinolates and their breakdown products on necrotrophic fungi. PLoS ONE 8:e70771. https://doi.org/10.1371/journal.pone.0070771
Carlsson M, von Bothmer R, Merker A (2004) Screening and evaluation of resistance to downy mildew (Peronospora parasitica) and clubroot (Plasmodiophora brassicae) in genetic resources of Brassica oleracea. Hereditas 141:293–300. https://doi.org/10.1111/j.1601-5223.2004.01818.x
Carmona D, Lajeunesse MJ, Johnson MTJ (2011) Plant traits that predict resistance to herbivores. Funct Ecol 25:358–367. https://doi.org/10.1111/J.1365-2435.2010.01794.X@10.1111/(ISSN)1365-2435.INTERCOLL
Carputo D, Frusciante L, Peloquin SJ (2003) The role of 2n gametes and endosperm balance number in the origin and evolution of polyploids in the tuber-bearing Solanums. Genetics 163:287–294
Cartea ME, Francisco M, Lema M et al (2010) Resistance of Cabbage (Brassica oleracea capitata Group) crops to Mamestra brassicae. J Econ Entomol 103:1866–1874. https://doi.org/10.1603/EC09375
Chae E, Bomblies K, Kim ST et al (2014) Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159:1341–1351. https://doi.org/10.1016/j.cell.2014.10.049
Chalhoub B, Denoeud F, Liu S et al (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953. https://doi.org/10.1126/science.1253435
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103:551–560. https://doi.org/10.1093/aob/mcn125
Chen S, Nelson MN, Chèvre AM et al (2011) Trigenomic bridges for Brassica improvement. CRC Crit Rev Plant Sci 30:524–547. https://doi.org/10.1080/07352689.2011.615700
Chen S, Wan ZJ, Nelson MN et al (2013) Evidence from genome-wide simple sequence repeat markers for a polyphyletic origin and secondary centers of genetic diversity of Brassica juncea in China and India. J Hered 104:416–427. https://doi.org/10.1093/jhered/est015
Cheng F, Mandakova T, Wu J et al (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25:1541–1554. https://doi.org/10.1105/tpc.113.110486
Cheng F, Sun R, Hou X et al (2016) Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet 48:1218–1224. https://doi.org/10.1038/ng.3634
Cheng L, Abraham J, Zhu J et al (2020) Record-setting ocean warmth continued in 2019. Adv Atmos Sci 37:137–142. https://doi.org/10.1007/s00376-020-9283-7
Choi K, Henderson IR (2015) Meiotic recombination hotspots: a comparative view. Plant J 83:52–61. https://doi.org/10.1111/tpj.12870
Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90:856–867. https://doi.org/10.1111/tpj.13299
Chu M, Yu F, Falk KC et al (2013) Identification of the clubroot resistance gene Rpb1 and introgression of the resistance into canola breeding lines using a marker-assisted approach. Acta Hortic 1005:599–605
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland
Darrier B, Rimbert H, Balfour F et al (2017) High-resolution mapping of crossover events in the hexaploid wheat genome suggests a universal recombination mechanism. Genetics 206(3):1373–1388
Delourme R, Chèvre AM, Brun H et al (2006) Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus). Eur J Plant Pathol 114:41–52. https://doi.org/10.1007/s10658-005-2108-9
Dhooghe E, van Laere K, Eeckhaut T et al (2011) Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tissue Organ Cult 104:359–373. https://doi.org/10.1007/s11240-010-9786-5
Dixon GR (2007) Vegetable Brassicas and related crucifers. In: Atherton J, Rees H (eds) Crop production science in horticulture series. CAB International
Dixon GR (2009) The occurrence and economic impact of Plasmodiophora brassicae and Clubroot Disease. J Plant Growth Regul 28:194–202. https://doi.org/10.1007/s00344-009-9090-y
Dosdall LM (2009) Responses of the cabbage seedpod weevil, Ceutorhynchus obstrictus (Marsham) (Coleoptera: Curculionidae), to seed treatments of canola (Brassica napus L.) with the neonicotinoid compounds clothianidin and imidacloprid. Pest Manag Sci 65:1329–1336. https://doi.org/10.1002/ps.1818
Eckardt NA (2006) Cytoplasmic male sterility and fertility restoration. Plant Cell 18:515–517. https://doi.org/10.1105/tpc.106.041830
Evivie ER, Ogwu MC, Cang W et al (2019) Progress and prospects of glucosinolate pathogen resistance in some brassica plants. J Appl Nat Sci 11:556–567. https://doi.org/10.31018/jans.v11i2.2117
Fahey JW (2003) BRASSICAS. In: Caballero B (ed) Encyclopedia of food sciences and nutrition, 2nd edn. Academic Press, pp 606–615
Fang Z, Sun P, Liu Y et al (1997) A male sterile line with dominant gene (Ms) in cabbage (Brassica oleracea var. capitata) and its utilization for hybrid seed production. Euphytica 97:265–268. https://doi.org/10.1023/A:1003026523150
Ferdous MJ, Hossain MR, Park JI et al (2020) In-silico identification and differential expressions of LepR4-syntenic disease resistance related domain containing genes against blackleg causal fungus Leptosphaeria maculans in Brassica oleracea. Gene Reports 19:100598. https://doi.org/10.1016/j.genrep.2020.100598
FitzJohn RG, Armstrong TT, Newstrom-Lloyd LE et al (2007) Hybridisation within Brassica and allied genera: evaluation of potential for transgene escape. Euphytica 158:209–230
Fletcher RS, Herrmann D, Mullen JL et al (2016) Identification of polymorphisms associated with drought adaptation QTL in Brassica napus by resequencing. G3 Genes Genomes. Genet 6:793–803. https://doi.org/10.1534/g3.115.021279
Fletcher RS, Mullen JL, Heiliger A, McKay JK (2015) QTL analysis of root morphology, flowering time, and yield reveals trade-offs in response to drought in Brassica napus. J Exp Bot 66:245–256. https://doi.org/10.1093/jxb/eru423
Frandsen KJ (1947) The experimental formation of Brassica napus L. var. oleifera DC. and Brassica carinata Braun. Dansk Bot Ark 12:1–16
Fredua-Agyeman R, Hwang SF, Strelkov SE et al (2019) Identification of Brassica accessions resistant to ‘old’ and ‘new’ pathotypes of Plasmodiophora brassicae from Canada. Plant Pathol 68:708–718. https://doi.org/10.1111/ppa.12980
Friedt W, Tu J, Fu T (2018) Academic and economic importance of Brassica napus rapeseed. In: Liu S, Snowdon R, Chalhoub B (eds) The Brassica napus Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-319-43694-4_1
Gaebelein R, Alnajar D, Koopmann B, Mason AS (2019) Hybrids between Brassica napus and B. nigra show frequent pairing between the B and A/C genomes and resistance to blackleg. Chromosom Res. https://doi.org/10.1007/s10577-019-09612-2
Gaebelein R, Mason AS (2018) Allohexaploids in the genus Brassica. CRC Crit Rev Plant Sci 37:422–437. https://doi.org/10.1080/07352689.2018.1517143
Gill SS, Khan NA, Tuteja N (2011) Differential cadmium stress tolerance in five Indian mustard (Brassica juncea L.) cultivars. Plant Signal Behav 6:293–300. https://doi.org/10.4161/psb.6.2.15049
Gomez-Campo C (1980) Morphology and morphotaxonomy of the tribe Brassiceae. In: Tsunoda S, Hinata K, Gomez-Campo C (eds) Brassica crops and wild allies: biology and breeding. Japan Scientific Soc Press, pp 3–31
Grandont L, Cuñado N, Coriton O et al (2014) Homoeologous chromosome sorting and progression of meiotic recombination in Brassica napus: ploidy does matter! Plant Cell 26:1448–1463
Gunasinghe N, You MP, Banga SS et al (2017) Outstanding host resistance will resolve the threat from white leaf spot disease (Pseudocercosporella capsellae) to oilseed and vegetable Brassica spp.crops. Australas Plant Pathol 46:137–146. https://doi.org/10.1007/s13313-017-0470-7
Gunasinghe N, You MP, Banga SS, Barbetti MJ (2014) High level resistance to Pseudocercosporella capsellae offers new opportunities to deploy host resistance to effectively manage white leaf spot disease across major cruciferous crops. Eur J Plant Pathol 138:873–890. https://doi.org/10.1007/s10658-013-0360-y
Gunasinghe N, You MP, Clode PL, Barbetti MJ (2016) Mechanisms of resistance in Brassica carinata, B. napus and B. juncea to Pseudocercosporella capsellae. Plant Pathol 65:888–900. https://doi.org/10.1111/ppa.12484
Haig D, Westoby M (1991) Seed size, pollination costs and angiosperm success. Evol Ecol 5:231–247. https://doi.org/10.1007/BF02214230
Hansen LM (2004) Economic damage threshold model for pollen beetles (Meligethes aeneus F.) in spring oilseed rape (Brassica napus L.) crops. Crop Prot 23:43–46. https://doi.org/10.1016/S0261-2194(03)00167-4
Hayat S, Mir BA, Wani AS et al (2011) Screening of salt-tolerant genotypes of Brassica juncea based on photosynthetic attributes. J Plant Interact 6:53–60. https://doi.org/10.1080/17429145.2010.521592
Hernández-Allica J, Becerril JM, Garbisu C (2008) Assessment of the phytoextraction potential of high biomass crop plants. Environ Pollut 152:32–40. https://doi.org/10.1016/J.ENVPOL.2007.06.002
Hervé MR (2018) Breeding for insect resistance in oilseed rape: challenges, current knowledge and perspectives. Plant Breed 137:27–34. https://doi.org/10.1111/pbr.12552
Hervé MR, Cortesero AM (2016) Potential for oilseed rape resistance in pollen beetle control. Arthropod Plant Interact 10:463–475. https://doi.org/10.1007/s11829-016-9438-8
Heyn FJW (1977) Analysis of unreduced gametes in Brassiceae by crosses between species and ploidy levels. J Plant Breed 78:13–30
Hopkins RJ, Griffiths DW, McKinlay RG, Birch ANE (1999) The relationship between cabbage root fly (Delia radicum) larval feeding and the freeze-dried matter and sugar content of Brassica roots. Entomol Exp Appl 92:109–117. https://doi.org/10.1046/j.1570-7458.1999.00530.x
Huangfu C, Song X, Qiang S (2009) Morphological disparities in the epidermal and anatomical features of the leaf among wild Brassica juncea populations. Weed Biol Manag 9:234–242. https://doi.org/10.1111/j.1445-6664.2009.00344.x
Iniguez-Luy FL, Federico ML (2011) The genetics of Brassica napus. In: Schmidt R, Bancroft I (eds) Genetics and Genomics of the Brassicaceae. Springer, pp 291–322
IPCC (2014) Climate change 2014, synthesis report of the IPCC fifth assessment report (AR5)
IPCC (2018) Global warming of 1.5°C. Summary for policymakers
Irfan M, Ahmad A, Hayat S (2014) Effect of cadmium on the growth and antioxidant enzymes in two varieties of Brassica juncea. Saudi J Biol Sci 21:125–131. https://doi.org/10.1016/J.SJBS.2013.08.001
Iwaya-Inoue M, Sakurai M, Uemura M (2018) Survival strategies in extreme cold and desiccation. Springer, Singapore
Jenczewski E, Eber F, Grimaud A et al (2003) PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164:645–653
Jestin C, Lodé M, Vallée P et al (2011) Association mapping of quantitative resistance for Leptosphaeria maculans in oilseed rape (Brassica napus L.). Mol Breed 27:271–287. https://doi.org/10.1007/s11032-010-9429-x
Johnson NA (2010) Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet 26:317–325. https://doi.org/10.1016/j.tig.2010.04.005
Karling JS, Karling JS (1942) The Plasmodiophorales; including a complete host index, bibliography, and a description of diseases caused by species of this order, 1st edn. The author, New York city
Katche E, Quezada-Martinez D, Katche EI et al (2019) Interspecific hybridization for Brassica crop improvement. Crop Breed Genet Genom 1:e190007
Katz E, Nisani S, Yadav BS et al (2015) The glucosinolate breakdown product indole-3-carbinol acts as an auxin antagonist in roots of Arabidopsis thaliana. Plant J 82:547–555. https://doi.org/10.1111/tpj.12824
Kim JK, Choi SR, Lee J et al (2013) Metabolic differentiation of diamondback moth (Plutella xylostella (L.)) resistance in cabbage (Brassica oleracea L. ssp. capitata). J Agric Food Chem 61:11222–11230. https://doi.org/10.1021/jf403441t
Koch S, Dunker S, Kleinhenz B et al (2007) A crop loss-related forecasting model for sclerotinia stem rot in winter oilseed rape. Phytopathology 97:1186–1194. https://doi.org/10.1094/PHYTO-97-9-1186
Kodra E, Bhatia U, Chatterjee S et al (2020) Physics-guided probabilistic modeling of extreme precipitation under climate change. Sci Rep 10:10299. https://doi.org/10.1038/s41598-020-67088-1
Köhler C, Mittelsten Scheid O, Erilova A et al (2010) The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet 26:142–148. https://doi.org/10.1016/j.tig.2009.12.006
Kumar G, Purty RS, Sharma MP et al (2009) Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J Plant Physiol 166:507–520. https://doi.org/10.1016/J.JPLPH.2008.08.001
Kumar S, Sangha MK (2017) Biochemical mechanism of resistance in some Brassica genotypes against Lipaphis erysimi (Kaltenbach) (Homoptera: Aphididae). Vegetos Int J Plant Res 26:387–395
Lafon-Placette C, Köhler C (2016) Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Mol Ecol 25:2620–2629. https://doi.org/10.1111/mec.13552
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144:1903–1910. https://doi.org/10.1016/j.cbd.2009.01.003
Larkan NJ, Raman H, Lydiate DJ et al (2016) Multi-environment QTL studies suggest a role for cysteine-rich protein kinase genes in quantitative resistance to blackleg disease in Brassica napus. BMC Plant Biol 16:1–16. https://doi.org/10.1186/s12870-016-0877-2
Lee RWH, Malchev IT, Rajcan I, Kott LS (2014) Identification of putative quantitative trait loci associated with a flavonoid related to resistance to cabbage seedpod weevil (Ceutorhynchus obstrictus) in canola derived from an intergeneric cross, Sinapis alba × Brassica napus. Theor Appl Genet 127:419–428. https://doi.org/10.1007/s00122-013-2228-0
Leflon M, Brun H, Eber F et al (2007) Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus. Theor Appl Genet 115:897–906. https://doi.org/10.1007/s00122-007-0616-z
Leflon M, Grandont L, Eber F et al (2010) Crossovers get a boost in Brassica allotriploid and allotetraploid hybrids. Plant Cell 22:2253–2264
Lema M, Cartea ME, Francisco M et al (2015) Screening for resistance to black rot in a Spanish collection of Brassica rapa. Plant Breed 134:551–556. https://doi.org/10.1111/pbr.12293
Lema M, Velasco P, Soengas P et al (2012) Screening for resistance to black rot in Brassica oleracea crops. Plant Breed 131:607–613. https://doi.org/10.1111/j.1439-0523.2012.01974.x
Li CX, Liu SY, Sivasithamparam K, Barbetti MJ (2009) New sources of resistance to Sclerotinia stem rot caused by Sclerotinia sclerotiorum in Chinese and Australian Brassica napus and B. juncea germplasm screened under Western Australian conditions. Australas Plant Pathol 38:149–152. https://doi.org/10.1071/Ap08087
Li Z, Feng X, Liu S-S et al (2016) Biology, ecology, and management of the diamondback moth in China. Annu Rev Entomol 61:277–296. https://doi.org/10.1146/annurev-ento-010715-023622
Light KA, Gororo NN, Salisbury PA (2011) Usefulness of winter canola (Brassica napus) race-specific resistance genes against blackleg (causal agent Leptosphaeria maculans) in southern Australian growing conditions. Crop Pasture Sci 62:162–168. https://doi.org/10.1071/CP10187
Liu Z, Adamczyk K, Manzanares-Dauleux M et al (2006) Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174:1583–1596. https://doi.org/10.1534/genetics.106.064071
Lu J, Carbone GJ, Grego JM (2019) Uncertainty and hotspots in 21st century projections of agricultural drought from CMIP5 models. Sci Rep 9:157. https://doi.org/10.1038/s41598-019-41196-z
Luo Z, Wang M, Long Y et al (2017) Incorporating pleiotropic quantitative trait loci in dissection of complex traits: seed yield in rapeseed as an example. Theor Appl Genet. https://doi.org/10.1007/s00122-017-2911-7
Mageney V, Baldermann S, Albach DC (2016) Intraspecific variation in carotenoids of Brassica oleracea var. sabellica. J Agric Food Chem 64:3251–3257. https://doi.org/10.1021/acs.jafc.6b00268
Manzanares-Dauleux MJ, Delourme R, Baron F, Thomas G (2000) Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theor Appl Genet 101:885–891. https://doi.org/10.1007/s001220051557
Maskell LC, Raybould AF, Cooper JI et al (1999) Effects of turnip mosaic virus and turnip yellow mosaic virus on the survival, growth and reproduction of wild cabbage (Brassica oleracea). Ann Appl Biol 135:401–407. https://doi.org/10.1111/j.1744-7348.1999.tb00867.x
Mason AS, Batley J, Bayer PE et al (2014) High-resolution molecular karyotyping uncovers pairing between ancestrally related Brassica chromosomes. New Phytol 202:964–974. https://doi.org/10.1111/nph.12706
Mason AS, Chèvre AM (2016) Optimization of recombination in interspecific hybrids to introduce new genetic diversity into oilseed rape (Brassica napus L.). In: Mason AS (ed) Polyploidy and hybridization for crop improvement. CRC Press, Boca Raton, USA, pp 431–444
Mason AS, Huteau V, Eber F et al (2010) Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosom Res 18:655–666. https://doi.org/10.1007/s10577-010-9140-0
Mason AS, Snowdon RJ (2016) Oilseed rape: learning about ancient and recent polyploid evolution from a recent crop species. Plant Biol 18:883–892
Mei J, Ding Y, Lu K et al (2013) Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor Appl Genet 126:549–556. https://doi.org/10.1007/s00122-012-2000-x
Mei J, Liu Y, Wei D et al (2015) Transfer of sclerotinia resistance from wild relative of Brassica oleracea into Brassica napus using a hexaploidy step. Theor Appl Genet 128:639–644. https://doi.org/10.1007/s00122-015-2459-3
Mei J, Qian L, Disi JO et al (2011) Identification of resistant sources against Sclerotinia sclerotiorum in Brassica species with emphasis on B. oleracea. Euphytica 177:393–399. https://doi.org/10.1007/s10681-010-0274-0
Mei J, Shao C, Yang R et al (2020) Introgression and pyramiding of genetic loci from wild Brassica oleracea into B. napus for improving Sclerotinia resistance of rapeseed. Theor Appl Genet 133:1313–1319. https://doi.org/10.1007/s00122-020-03552-w
Meng H, Jiang S, Hua S et al (2011) Comparison between a tetraploid turnip and its diploid progenitor (Brassica rapa L.): The adaptation to salinity stress. Agric Sci China 10:363–375. https://doi.org/10.1016/S1671-2927(11)60015-1
Mizushima U (1980) Genome analysis in Brassica and allied genera. In: Tsunoda S, Hinata K G-CC (ed) Brassica crops and wild allies: biology and breeding. Japan Scientific Societies Press, Tokyo, pp 89–106
Monot C, Silué D (2009) Comparison of the resistance and susceptibility to downy mildew [Hyaloperonospora parasitica, constant. (Pers. Ex Fr)] of nine Brassica olearacea accessions in laboratory, seedbed, and field screens. Euphytica 169:413–419. https://doi.org/10.1007/s10681-009-9974-8
Mourato M, Moreira I, Leitão I et al (2015) Effect of heavy metals in plants of the genus Brassica. Int J Mol Sci 16:17975–17998. https://doi.org/10.3390/ijms160817975
Navabi ZK, Strelkov SE, Good AG et al (2010) Brassica B-genome resistance to stem rot (Sclerotinia sclerotiorum) in a doubled haploid population of Brassica napus × Brassica carinata. Can J Plant Pathol 32:237–246. https://doi.org/10.1080/07060661.2010.484229
Navrátilová B (2004) Protoplast cultures and protoplast fusion focused on Brassicaceae - a review. Hortic Sci 31:140–157. https://doi.org/https://doi.org/10.17221/3809-hortsci
Nerem RS, Beckley BD, Fasullo JT et al (2018) Climate-change–driven accelerated sea-level rise detected in the altimeter era. Proc Natl Acad Sci U S A 115:2022–2025. https://doi.org/10.1073/pnas.1717312115
Nicolas SD, Le Mignon G, Eber F et al (2007) Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175:487–503. https://doi.org/10.1534/genetics.106.062968
Nicolas SD, Leflon M, Monod H et al (2009) Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. Plant Cell Online 21:373–385. https://doi.org/10.1105/tpc.108.062273
Nishi S (1980) Differentiation of Brassica crops in Asia and the breeding of “Hakuran”, a newly synthesized leafy vegetable. In: Tsunoda S, Hinata KG-CC (eds) Brassica crops and wild allies. Japan Scientific Societies Press, Tokyo, pp 133–150
Nishiyama I, Sarashima M, Matsuzawa Y (1991) Critical discussion on abortive interspecific crosses in Brassica. Plant Breed 107:288–302. https://doi.org/10.1111/j.1439-0523.1991.tb00552.x
Nyalugwe EP, Barbetti MJ, Jones RAC (2016) Strain specificity of Turnip mosaic virus resistance gene TuRBJU01 in Brassica juncea. Eur J Plant Pathol 145:209–213. https://doi.org/10.1007/s10658-015-0816-3
Nyalugwe EP, Barbetti MJ, Jones RAC (2015) Studies on resistance phenotypes to Turnip mosaic virus in five species of Brassicaceae, and identification of a virus resistance gene in Brassica juncea. Eur J Plant Pathol 141:647–666. https://doi.org/10.1007/s10658-014-0568-5
Nyalugwe EP, Barbetti MJ, Jones RAC (2014) Preliminary studies on resistance phenotypes to Turnip mosaic virus in Brassica napus and B. carinata from different continents and effects of temperature on their expression. Eur J Plant Pathol 139:687–706. https://doi.org/10.1007/s10658-014-0423-8
Oduor AMO, Gómez JM, Herrador MB, Perfectti F (2015) Invasion of Brassica nigra in North America: Distributions and origins of chloroplast dna haplotypes suggest multiple introductions. Biol Invasions 17:2447–2459. https://doi.org/10.1007/s10530-015-0888-1
Ogura H (1968) Studies on the new male-sterility in Japanese radish, with special reference to the utilization of this sterility towards the practical raising of hybrid seeds. Mem Fac Agric Kagoshima Univ 6:39–78
Okamoto A, Ureshino K (2015) Pre- and post-fertilization barriers in interspecific hybridization between evergreen azalea species and Rhododendron uwaense H. Hara & T Yamanaka Hortic J 84:355–364. https://doi.org/10.2503/hortj.MI-036
Panjabi-Massand P, Yadava SK, Sharma P et al (2010) Molecular mapping reveals two independent loci conferring resistance to Albugo candida in the east European germplasm of oilseed mustard Brassica juncea. Theor Appl Genet 121:137–145. https://doi.org/10.1007/s00122-010-1297-6
Parkin IAP, Koh C, Tang H et al (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15:R77. https://doi.org/10.1186/gb-2014-15-6-r77
Parkin IAP, Sharpe AG, Lydiate DJ (2003) Patterns of genome duplication within the Brassica napus genome. Genome 46:291–303. https://doi.org/10.1139/G03-006
Pelé A, Trotoux G, Eber F et al (2017) The poor lonesome A subgenome of Brassica napus var. Darmor (AACC) may not survive without its mate. New Phytol 213:1886–1897. https://doi.org/10.1111/nph.14147
Pelgrom KTB, Broekgaarden C, Voorrips RE et al (2015) Host plant resistance towards the cabbage whitefly in Brassica oleracea and its wild relatives. Euphytica 202:297–306. https://doi.org/10.1007/s10681-014-1306-y
Pellan-Delourme R, Renard M (1988) Cytoplasmic male sterility in rapeseed (Brassica napus L.): female fertility of restored rapeseed with “Ogura” and cybrids cytoplasms. Genome 30:234–238
Pelletier G, Primard C, Vedel F et al (1983) Intergeneric cytoplasmic hybridization in cruciferae by protoplast fusion. MGG Mol Gen Genet 191:244–250. https://doi.org/10.1007/BF00334821
Peng G, Falk KC, Gugel RK et al (2014) Sources of resistance to Plasmodiophora brassicae (clubroot) pathotypes virulent on canola. Can J Plant Pathol 36:89–99. https://doi.org/10.1080/07060661.2013.863805
Peng Y, Shi D, Zhang T et al (2015) Development and utilization of an efficient cytoplasmic male sterile system for Cai-xin (Brassica rapa L.). Sci Hortic (Amsterdam) 190:36–42. https://doi.org/10.1016/J.SCIENTA.2015.04.002
Piao Z, Ramchiary N, Lim YP (2009) Genetics of clubroot resistance in Brassica species. J Plant Growth Regul 28:252–264. https://doi.org/10.1007/s00344-009-9093-8
Pilet-Nayel ML, Moury B, Caffier V et al (2017) Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front Plant Sci 8:27
Potts BM, Dungey HS (2004) Interspecific hybridization of Eucalyptus: key issues for breeders and geneticists. New For 27:115–138. https://doi.org/10.1023/a:1025021324564
Prabha K, Sood R, Gupta SC (1982) High temperature-induced inactivation of sporophytic self-incompatibility in Ipomoea fistula. New Phytol 92:115–122. https://doi.org/10.1111/j.1469-8137.1982.tb03367.x
Prakash S, Bhat SR, Quiros CF et al (2009) Brassica and its close allies: cytogenetics and evolution. In: Janick J (ed) Plant Breeding Reviews. John Wiley & Sons Inc, Hoboken, NJ, USA, pp 21–187
Price JS, Hobson RN, Neale MA, Bruce DM (1996) Seed losses in commercial harvesting of oilseed rape. J Agric Eng Res 65:183–191. https://doi.org/10.1006/jaer.1996.0091
Primard-Brisset C, Poupard JP, Horvais R et al (2005) A new recombined double low restorer line for the Ogu-INRA cms in rapeseed (Brassica napus L.). Theor Appl Genet 111:736–746. https://doi.org/10.1007/s00122-005-2059-8
Princen LH (1979) New crop developments for industrial oils. J Am Oil Chem Soc 56:845–848. https://doi.org/10.1007/BF02909532
Rahman M, Mamidi S, del Rio L et al (2016) Association mapping in Brassica napus (L.) accessions identifies a major QTL for blackleg disease resistance on chromosome A01. Mol Breed 36:1–15. https://doi.org/10.1007/s11032-016-0513-8
Rakow G (2004) Species origin and economic importance of Brassica. In: Pua EC, Douglas CJ (eds) Brassica. Biotechnology in Agriculture and Forestry, Springer, Berlin, Heidelberg, pp 3–11
Raman H, Raman R, Coombes N et al (2016) Genome-wide association study identifies new loci for resistance to Leptosphaeria maculans in canola. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01513
Raman H, Raman R, Kilian A et al (2014) Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS ONE 9:e101673. https://doi.org/10.1371/journal.pone.0101673
Rana K, Atri C, Gupta M et al (2017) Mapping resistance responses to Sclerotinia infestation in introgression lines of Brassica juncea carrying genomic segments from wild Brassicaceae B. fruticulosa. Sci Rep. https://doi.org/10.1038/s41598-017-05992-9
Raybould AF, Maskell LC, Edwards M-L et al (1999) The prevalence and spatial distribution of viruses in natural populations of Brassica oleracea. New Phytol 141:265–275
Ribeiro da Silva AL, Secchi Candian J, Ramalho do Rego E, et al (2020) Screening cabbage cultivars for resistance to black rot under field conditions. Am Soc Hortic Sci 30:448–455
Rieseberg LH, Blackman BK (2010) Speciation genes in plants. Ann Bot 106:439–455. https://doi.org/10.1093/aob/mcq126
Rimmer SR (2006) Resistance genes to Leptosphaeria maculans in Brassica napus. Can J Plant Pathol 28:S288–S297. https://doi.org/10.1080/07060660609507386
Saha P, Kalia P, Sharma M, Singh D (2016) New source of black rot disease resistance in Brassica oleracea and genetic analysis of resistance. Euphytica 207:35–48. https://doi.org/10.1007/s10681-015-1524-y
Salt DE, Smith RD, Raskin I (1998) Phytoremediation. Annu Rev Plant Physiol Plant Mol Biol 49:643–668. https://doi.org/10.1146/annurev.arplant.49.1.643
Santolamazza-Carbone S, Velasco P, Cartea ME (2017) Resistance to the cabbage root fly, Delia radicum (Diptera, Anthomyiidae), of turnip varieties (Brassica rapa subsp. rapa). Euphytica 213:274. https://doi.org/10.1007/s10681-017-2069-z
Sasidharan R, Hartman S, Liu Z et al (2018) Signal dynamics and interactions during flooding stress. Plant Physiol 176:1106–1117. https://doi.org/10.1104/pp.17.01232
Schaefer-Koesterke HL, Brandes H, Ulber B et al (2017) The potential of resynthesized lines to provide resistance traits against rape stem weevil in oilseed rape. J Pest Sci 90:87–101. https://doi.org/10.1007/s10340-016-0742-y
Schiessl S, Iniguez-Luy F, Qian W, Snowdon RJ (2015) Diverse regulatory factors associate with flowering time and yield responses in winter-type Brassica napus. BMC Genomics 16:737
Schiessl S, Samans B, Huttel B et al (2014) Capturing sequence variation among flowering-time regulatory gene homologs in the allopolyploid crop species Brassica napus. Front Plant Sci 5:404. https://doi.org/10.3389/fpls.2014.00404
Schiessl SV, Quezada-Martinez D, Orantes-Bonilla M, Snowdon RJ (2020) Transcriptomics reveal high regulatory diversity of drought tolerance strategies in a biennial oil crop. Plant Sci 297:110515. https://doi.org/10.1016/j.plantsci.2020.110515
Scholze P, Krämer R, Ryschka U et al (2010) Somatic hybrids of vegetable brassicas as source for new resistances to fungal and virus diseases. Euphytica 176:1–14. https://doi.org/10.1007/s10681-010-0205-0
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542
Schultz ES (1921) A transmissible mosaic disease of chinese cabbage, mustard, and turnip. J Agric Res 22:173–177
Sharma DR, Kaur R, Kumar K (1996) Embryo rescue in plants - A review. Euphytica 89:325–337. https://doi.org/10.1007/BF00022289
Sharma BB, Kalia P, Yadava DK, et al (2016) Genetics and molecular mapping of black rot resistance locus Xca1bc on chromosome B7 in Ethiopian mustard (Brassica carinata A. Braun). PLoS One 11:e0152290. https://doi.org/10.1371/journal.pone.0152290
Sharma BB, Kalia P, Singh D, Sharma TR (2017) Introgression of black rot resistance from Brassica carinata to Cauliflower (Brassica oleracea botrytis Group) through embryo rescue. Front Plant Sci 8:1255. https://doi.org/10.3389/fpls.2017.01255
Shattuck VI (2010) The biology, epidemiology, and control of Turnip Mosaic Virus. In: Horticultural Reviews. John Wiley & Sons, Inc., pp 199–238
Shuhang W, Voorrips RE, Steenhuis-Broers G et al (2016) Antibiosis resistance against larval cabbage root fly, Delia radicum, in wild Brassica-species. Euphytica 211:139–155. https://doi.org/10.1007/s10681-016-1724-0
Shukla PR, Skea J, Slade R, et al (2019) Climate Change and Land. An IPCC Special Report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems. Technical Summary.
Sinclair TR, Rufty TW, Lewis RS (2019) Increasing photosynthesis: unlikely solution for world food problem. Trends Plant Sci 24:1032–1039. https://doi.org/10.1016/j.tplants.2019.07.008
Singh D, Dhar S, Yadava DK (2011) Genetic and pathogenic variability of Indian strains of Xanthomonas campestris pv. campestris causing black rot disease in crucifers. Curr Microbiol 63:551–560. https://doi.org/10.1007/s00284-011-0024-0
Snogerup S (1980) The wild forms of the Brassica oleracea group (2n = 18) and their possible relations to the cultivated ones. In: Tsunoda S, Hinata K, Gomez-Campo C (eds) Brassica crops and wild allies. Japan Scientific Societies Press, Tokyo, pp 121–132
Snowdon R (2007) Cytogenetics and genome analysis in Brassica crops. Chromosom Res 15:85–95. https://doi.org/10.1007/s10577-006-1105-y
Snowdon RJ, Luy FLI (2012) Potential to improve oilseed rape and canola breeding in the genomics era. Plant Breed 131:351–360. https://doi.org/10.1111/j.1439-0523.2012.01976.x
Spence NJ, Phiri NA, Hughes SL et al (2007) Economic impact of Turnip mosaic virus, Cauliflower mosaic virus and Beet mosaic virus in three Kenyan vegetables. Plant Pathol 56:317–323. https://doi.org/10.1111/j.1365-3059.2006.01498.x
Spitzer T, Bílovský J, Matušinsky P (2020) Changes in resistance development in pollen beetle (Brassicogethes aeneus F.) to lambda-cyhalothrin, etofenprox, chlorpyrifos-ethyl, and thiacloprid in the Czech Republic during 2013–2017. Crop Prot 135:105224. https://doi.org/10.1016/j.cropro.2020.105224
Sun B, Tian Y-X, Chen Q et al (2019) Variations in the glucosinolates of the individual edible parts of three stem mustards (Brassica juncea ). R Soc Open Sci 6:182054. https://doi.org/10.1098/rsos.182054
Suzuki N, Rivero RM, Shulaev V et al (2014) Abiotic and biotic stress combinations. New Phytol 203:32–43. https://doi.org/10.1111/nph.12797
Świadek M, Proost S, Sieh D et al (2017) Novel allelic variants in ACD6 cause hybrid necrosis in local collection of Arabidopsis thaliana. New Phytol 213:900–915. https://doi.org/10.1111/nph.14155
Talbert PB, Henikoff S (2010) Centromeres convert but don’t cross. PLoS Biol 8:e1000326. https://doi.org/10.1371/journal.pbio.1000326
Tansey JA, Dosdall LM, Keddie A et al (2010) Antixenosis and antibiosis resistance to Ceutorhynchus obstrictus in novel germplasm derived from Sinapis alba x Brassica napus. Can Entomol 142:212–221. https://doi.org/10.4039/n09-067
Tateda C, Zhang Z, Greenberg JT (2015) Linking pattern recognition and salicylic acid responses in arabidopsis through ACCELERATED CELL DEATH6 and receptors. Plant Signal Behav 10:e1010912. https://doi.org/10.1080/15592324.2015.1010912
Taylor A, Coventry E, Jones JE, Clarkson JP (2015) Resistance to a highly aggressive isolate of Sclerotinia sclerotiorum in a Brassica napus diversity set. Plant Pathol 64:932–940. https://doi.org/10.1111/ppa.12327
The Brassica rapa Genome Sequencing Project Consortium (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039. https://doi.org/10.1038/Ng.919
Tian E, Roslinsky V, Cheng B (2014) Molecular marker-assisted breeding for improved Ogura cms restorer line (RfoRfo) and mapping of the restorer gene (Rfo) in Brassica juncea. Mol Breed. https://doi.org/10.1007/s11032-014-0121-4
Todesco M, Kim ST, Chae E et al (2014) Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. PLoS Genet 10:e1004459. https://doi.org/10.1371/journal.pgen.1004459
Traka M, Mithen R (2009) Glucosinolates, isothiocyanates and human health. Phytochem Rev 8:269–282. https://doi.org/10.1007/s11101-008-9103-7
Trenberth KE (2011) Changes in precipitation with climate change. Clim Res 47:123–138. https://doi.org/10.3354/cr00953
Tsunoda S (1980) Eco-physiology of wild and cultivated forms in Brassica and allied genera. In: Tsunoda S, Hinata K, Gómez-Campo C (eds) Brassica crops and wild allies. Japan Scientific Societies Press, Sendai, Japan, pp 109–120
U N, (1935) Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japanese J Bot 7:389–452
Ureshino K, Miyajima I, Ozaki Y et al (1999) Appearance of albino seedlings and ptDNA inheritance in interspecific hybrids of azalea. Euphytica 110:61. https://doi.org/10.1023/A:1003754004737
Vaid N, Laitinen RAE (2019) Diverse paths to hybrid incompatibility in Arabidopsis. Plant J 97:199–213. https://doi.org/10.1111/tpj.14061
Vaughan JG (1977) A multidisciplinary study of the taxonomy and origin of Brassica crops. Bioscience 27:35–40. https://doi.org/10.2307/1297791
Verkerk R, Tebbenhoff S, Dekker M (2010) Variation and distribution of glucosinolates in 42 cultivars of Brassica oleracea vegetable crops. Acta Hortic. https://doi.org/10.17660/ActaHortic.2010.856.7
Vicente JG, Conway J, Roberts SJ, Taylor JD (2001) Identification and origin of Xanthomonas campestris pv. campestris Races and related pathovars. Phytopathology 91:492–499. https://doi.org/10.1094/PHYTO.2001.91.5.492
Walsh JA, Jenner CE (2002) Turnip mosaic virus and the quest for durable resistance. Mol Plant Pathol 3:289–300. https://doi.org/10.1046/j.1364-3703.2002.00132.x
Walsh JA, Sharpe AG, Jenner CE, Lydiate DJ (1999) Characterisation of resistance to turnip mosaic virus in oilseed rape (Brassica napus) and genetic mapping of TuRB01. Theor Appl Genet 99:1149–1154. https://doi.org/10.1007/s001220051319
Wan Z, Jing B, Tu J et al (2008) Genetic characterization of a new cytoplasmic male sterility system (hau) in Brassica juncea and its transfer to B. napus. Theor Appl Genet 116:355–362. https://doi.org/10.1007/s00122-007-0673-3
Wang W, Guan R, Liu X et al (2019) Chromosome level comparative analysis of Brassica genomes. Plant Mol Biol 99:237–249. https://doi.org/10.1007/s11103-018-0814-x
Warwick S, Anderson J (1993) Guide to the wild germplasm of Brassica and allied crops: Part II: Chromosome numbers in the tribe Brassiceae (Cruciferae). Agric Canada Res Branch, Tech Bull 15E:22
Warwick SI, Francis A, Al-Shehbaz IA (2006) Brassicaceae: Species checklist and database on CD-Rom. Plant Syst Evol 259:249–258. https://doi.org/10.1007/s00606-006-0422-0
Warwick SI, Francis A, Gugel RK (2009) Part IV: Wild Crucifer Species as Sources of Agronomic Traits. Guide to Wild Germplasm Brassica Allied Crop (Tribe Brassiceae, Brassiceae), pp 1–60
Wei WL, Wang HZ, Liu GH (2009) Cytological and molecular characterization of a new cytoplasmic male sterility in rapeseed. Plant Breed 128:426–428. https://doi.org/10.1111/j.1439-0523.2008.01592.x
Wen Y-C (2008) Screening and analysis of resistance to silique shattering in rape (Brassica napus L.). Acta Agron Sin 34:163–166. https://doi.org/10.3724/SP.J.1006.2008.00163
West JS, Kharbanda PD, Barbetti MJ, Fitt BDL (2001) Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol 50:10–27. https://doi.org/10.1046/j.1365-3059.2001.00546.x
Wilson RA, Sangha MK, Banga SS et al (2014) Heat stress tolerance in relation to oxidative stress and antioxidants in Brassica juncea. J Environ Biol 35:383–387
Wu D, Liang Z, Yan T et al (2019) Whole-genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol Plant 12:30–43. https://doi.org/10.1016/j.molp.2018.11.007
Xiao Y, Chen L, Zou J et al (2010) Development of a population for substantial new type Brassica napus diversified at both A/C genomes. Theor Appl Genet 121:1141–1150. https://doi.org/10.1007/s00122-010-1378-6
Xiong Z-T, Wang H (2005) Copper toxicity and bioaccumulation in Chinese cabbage (Brassica pekinensis Rupr.). Environ Toxicol 20:188–194. https://doi.org/10.1002/tox.20094
Yamagishi H, Bhat SR (2014) Cytoplasmic male sterility in Brassicaceae crops. Breed Sci 64:38–47. https://doi.org/10.1270/jsbbs.64.38
Yang J, Liu D, Wang X et al (2016) The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet 48:1225–1232
Yangjun Y, Guang C, Jiabing X et al (2005) A new chinese cabbage F1 with orange color “Beijing Juhong 2.” Acta Hortic Sin 32:372–372
Yong H-Y, Wang C, Bancroft I et al (2015) Identification of a gene controlling variation in the salt tolerance of rapeseed (Brassica napus L.). Planta 242:313–326. https://doi.org/10.1007/s00425-015-2310-8
Yu F, Lydiate DJ, Rimmer SR (2005) Identification of two novel genes for blackleg resistance in Brassica napus. Theor Appl Genet 110:969–979. https://doi.org/10.1007/s00122-004-1919-y
Yu F, Lydiate DJ, Rimmer SR (2008) Identification and mapping of a third blackleg resistance locus in Brassica napus derived from B. rapa subsp. sylvestris. Genome 51:64–72. https://doi.org/10.1139/G07-103
Zalucki MP, Shabbir A, Silva R et al (2012) Estimating the economic cost of one of the world’s major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): Just How Long Is a Piece of String? J Econ Entomol 105:1115–1129. https://doi.org/10.1603/EC12107
Zhang F, Batley J (2020) Exploring the application of wild species for crop improvement in a changing climate. Curr Opin Plant Biol. https://doi.org/10.1016/j.pbi.2019.12.013
Zhang H, Feng J, Zhang S et al (2015a) Resistance to Plasmodiophora brassicae in Brassica rapa and Brassica juncea genotypes from China. Plant Dis 99:776–779. https://doi.org/10.1094/PDIS-08-14-0863-RE
Zhang J, Mason AS, Wu J et al (2015b) Identification of putative candidate genes for water stress tolerance in canola (Brassica napus). Front Plant Sci 6:1058. https://doi.org/10.3389/fpls.2015.01058
Zhiyuan F, Peitian S, Yumei L et al (1995) Preliminary study on the inheritance of male sterility in cabbage line 79–399–438. Acta Hortic. https://doi.org/10.17660/ActaHortic.1995.402.68
Zhou Q-H, Fu D-H, Mason AS et al (2014) In silico integration of quantitative trait loci for seed yield and yield-related traits in Brassica napus. Mol Breed 33:881–894. https://doi.org/10.1007/s11032-013-0002-2
Zhou S, Kai H, Zha Z et al (2016) Subcellular distribution and chemical forms of thorium in Brassica juncea var. foliosa. J Environ Radioact 157:60–66. https://doi.org/10.1016/J.JENVRAD.2016.03.003
Zhu M, Monroe JG, Suhail Y et al (2016) Molecular and systems approaches towards drought-tolerant canola crops. New Phytol 210:1169–1189. https://doi.org/10.1111/nph.13866
Zhu Z, Zheng W, Zhang X (2011) Principal component analysis and comprehensive evaluation on morphological and agronomic traits of drought tolerance in rapeseed (Brassica napus). Sci Agric Sin 44:1775–1787. https://doi.org/10.3864/J.ISSN.0578-1752.2011.09.003
Žnidarčič D, Valič N, Trdan S (2008) Epicuticular wax content in the leaves of cabbage (Brassica oleracea L. var. capitata) as a mechanical barrier against three insect pests. Acta Agric Slov 91:361–370. https://doi.org/10.2478/v10014-008-0016-3
Zou J, Zhu J, Huang S et al (2010) Broadening the avenue of intersubgenomic heterosis in oilseed Brassica. Theor Appl Genet 120:283–290. https://doi.org/10.1007/s00122-009-1201-4
Funding
Open Access funding enabled and organized by Projekt DEAL. ASM is funded by Deutsche Forschungsgemeinschaft Emmy Noether Grant MA6473/1–1, DQM by DFG Sino-German Grant MA6473/3–1, and SVS by DFG Grant 405304351.
Author information
Authors and Affiliations
Contributions
All authors contributed to writing and critical revision of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Consent for publication
All authors have seen and approved this submission.
Additional information
Communicated by Brent Hulke.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Quezada-Martinez, D., Addo Nyarko, C.P., Schiessl, S.V. et al. Using wild relatives and related species to build climate resilience in Brassica crops. Theor Appl Genet 134, 1711–1728 (2021). https://doi.org/10.1007/s00122-021-03793-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03793-3