Abstract
The perennial Secale cereanum cultivar ‘Kriszta’ is an artificial hybrid of S. cereale and S. strictum ssp. anatolicum. From the cross between the wheat line Mv9kr1 and ‘Kriszta’, which aimed the transfer of beneficial traits from rye to wheat, numerous translocation lines have been produced. For the identification of the translocated chromosomes, the unambiguous differentiation between chromosome arms of ‘Kriszta’ is essential. The identification of its short chromosome arms using conventional FISH probes is easy, but because of their similar hybridization patterns, its long arms cannot be distinguished. The present study aimed to create the detailed karyotype of ‘Kriszta’, especially that of long arms, by both chromosome measurements and FISH using highly repetitive, as well as subtelomeric tandem repeat, and synthetic microsatellite DNA sequences. Our results indicate that the chromosome complement of ‘Kriszta’ is not a simple combination of the chromosomes of the parental rye species but is composed of rearranged chromosomes. It is also showed that an adequate pair-wise combination of the DNA sequences pSc119.2, pSc200, pSc250, and (AAC)5 makes it possible to identify any of the long arms of S. cereanum cv. Kriszta chromosomes. The future usability of the identified wheat- ‘Kriszta’ translocated chromosomes is also discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the 1960s, crosses between Secale cereale (cv. Várda) and S. montanum (syn. S. strictum) ssp. anatolicum were carried out in Hungary to combine favorable agronomic characteristics of these rye species (Kotvics 1970). From the artificial hybrids produced, three perennial rye (S. cereanum) cultivars have been developed: ‘Kriszta’ (Kruppa 2001) and ‘Perenne’ (Füle et al. 2005) registered in 1998, and ‘Gergő’ registered in 2009 (not published). ‘Kriszta’ having high dietary fiber and protein content as well as tolerance to frost and drought, and resistance to leaf rust, stem rust, yellow (stripe) rust, and powdery mildew is a valuable source of genes for broadening the genetic diversity of bread wheat. In 2001, the hexaploid winter wheat line Martonvásári 9 kr1 (Molnár-Láng et al. 1996) was crossed with ‘Kriszta’ rye to transfer its beneficial traits into wheat. Since then, numerous backcrossed and selfed progeny plants carrying wheat-rye chromosomal rearrangements have been produced. For their future usability in different pre-breeding processes (e.g. tests for biotic and abiotic stresses or quality traits), determination of the precise composition of the translocation chromosomes is essential. Fluorescence in situ hybridization (FISH) is a powerful molecular cytogenetic technique to visualize repetitive or single-gene target DNA sequences, which allows the identification (by specific hybridization patterns) of plant chromosomes (Leitch et al. 1991; Said et al. 2018). Using pair-wise combinations of the repetitive sequences pSc119.2 (Bedbrook et al. 1980; McIntyre et al. 1990), (GAA)7 (Pedersen and Langridge 1997) and Afa family (Nagaki et al. 1995), identification of all wheat chromosomes (chromosome arms) is possible (Schneider et al. 2003). In the case of rye, besides the pSc119.2, the synthetic oligonucleotide (AAC)5 (Cuadrado and Schwarzacher 1998) and the subtelomeric, multicopy tandem repeat DNA sequences pSc200 and pSc250 (members of the 350–480 bp and 610 bp sequence families, respectively) (Vershinin et al. 1995) are the most commonly used FISH probes, which have been applied, first of all, in genetic studies of the phylogenetic relationships among Secale species (Achrem et al. 2014; Zhou et al. 2010). For the identification of ‘Kriszta’ chromosomes, only pSc119.2 (Szakács et al. 2020) and the combination of pSc119.2 and (AAC)5 (Schneider et al. 2016) have been used so far.
FISH identification of the rye chromosomes in an entire chromosome complement is relatively easy because the chromosome-specific arm ratio of the individual chromosomes is observable. In the above-mentioned introgression lines, where a single rye chromosome arm (pair) is present among the wheat chromosomes, comparison of the arm lengths is not possible.
The present study aimed to set up a detailed karyotype for ‘Kriszta’ rye based on chromosome measurements and FISH, as well as to determine individual chromosome arm landmarks using subtelomeric tandem repeat and synthetic microsatellite DNA sequences, which enable us to unambiguously identify ‘Kriszta’ chromosome arms in the previously produced wheat-perennial rye introgression lines.
Materials and methods
Plant materials
The following rye species were used in the karyotyping and chromosome arm identification studies: perennial Secale cereanum (2n = 2x = 14; RR; cv. Kriszta), S. cereale (2n = 2x = 14; RR; cv. Várda) and S. strictum (C. Presl) C. Presl. ssp. anatolicum (Boiss.) K. Hammer (2n = 2x = 14; RR; accession R797) from the Genebank of IPK Gatersleben.
Preparation of metaphase chromosome spreads
Chromosome preparations were made according to the method described by Endo and Gill (1984) with minor modifications. The seeds of the three species were germinated at room temperature for 24 h, and, to achieve synchronous cell division, they were incubated at 4 °C for 48 h and then at 25 °C for 26 h. After excision, root tips were treated in ice-cold sterile water for 24 h and fixed in Carnoy's solution I (1 part glacial acetic acid and 3 parts absolute ethanol). Root tips were squashed on degreased microscope slides using a drop of 45% acetic acid. The coverslips were removed in liquid nitrogen, and the preparations were air-dried overnight and stored at − 20 °C until use.
Chromosome measurement and karyotyping
Karyotype analysis was done on 10 complete metaphase cells (20 chromosomes/1R-7R) of five different root tips of each genotype. Preparations with well-spread chromosomes carrying detectable primary and secondary constrictions were selected. Measurements, calculation of the short arm length (S), long arm length (L), total chromosome length (TL; S + L), arm ratio (AR; L/S), and centromeric index (CI; S/S + L), as well as the construction of idiograms, were performed with the use of DRAWID software version 0.26 (Kirov et al. 2017). Length values were given by the software in pixels. The relative chromosome length (RL) was also calculated by dividing the length of a particular chromosome by the sum of lengths of all chromosomes in the haploid set and expressed in percent.
Fluorescence in situ hybridization (FISH)
FISH karyotyping of the rye chromosomes was carried out by using highly repetitive heterochromatic sequences pSc119.2, pSc200 and pSc250, and 12 synthetic simple sequence repeat oligonucleotide (microsatellite) probes: (AC)n, (AG)n, (AAC)n, (ACG)n, (ACT)n, (AGG)n, (ATT)n, (CAC)n, (CAG)n, (CAT)n, (GAA)n and (GCC)n. The highly repetitive DNA sequences pSc119.2, pSc200 and pSc250 were amplified from the genomic DNA of S. cereale (Zhou et al. 2010), while the oligonucleotides were amplified from the genomic DNA of T. aestivum as described in Contento et al. (2005). The PCR-amplified DNA sequences were labeled with digoxigenin-11-dUTP (Roche Diagnostics, Mannheim, Germany) by nick translation following the manufacturer's instructions, except for pSc119.2, which was labeled with biotin-16-dUTP (Roche Diagnostics, Mannheim, Germany). Hybridizations were performed using probe pairs composed of the biotinylated pSc119.2 and one of the digoxigenin-labeled repetitive sequences. Hybridization mixtures (30 µL per slide) contained 50% v/v formamide, 2 × SSC (0.15 mol/L NaCl plus 0.015 mol/L sodium citrate), 10% w/v dextran sulphate, 1.4 µg salmon sperm DNA, 0.1% w/v sodium dodecyl sulphate, and two DNA probes (50 ng each). The probe and target chromosomal DNAs were denatured simultaneously on the microscope slides at 75 ºC for 6 min and hybridized overnight at 37 ºC. Signals of the biotin- and digoxigenin-labeled probes were detected using anti-digoxigenin-Rhodamine (Roche) and streptavidin-FITC (Roche), respectively. Slides were counterstained with 1 µg/mL 4’,6-diamidino-2-phenylindole (DAPI, Amersham, Germany), coverslipped with Vectashield antifade mounting medium (Vector Laboratories, Burlingame, USA), and examined under a Zeiss Axio Imager 2 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany). Images were captured with a Zeiss AxioCam MRm CCD camera (Carl Zeiss Microimaging GmbH, Jena, Germany) using AxioVision SE64 Rel. 4.9.1 software (Carl Zeiss Microimaging GmbH, Jena, Germany).
Statistical analysis
One-way ANOVA function and Tukey's post hoc test of the IBM SPSS Statistics 20.0 software (SPSS Inc., Chicago, IL, USA) were used to evaluate data from the chromosome measurements.
Results
Chromosome measurement and karyotyping
A summary of the karyological features of the studied genotypes is given in Tables 1 and 2. Comparison of the total chromosome lengths in ‘Várda’, R797, and ‘Kriszta’, respectively, did not show differences between chromosomes 1R-7R. However, within-species variability in the relative lengths was found in S. cereale and S. strictum ssp. anatolicum (Table 1). In ‘Várda’, chromosome 3R proved to be significantly shorter than 6R, while the rest of the chromosomes did not differ significantly from either 3R or 6R. In R797, 1R was found to be significantly shorter than 2R, 3R, and 4R, while chromosomes 5R, 6R, and 7R did not differ significantly from the previous ones. Interestingly, the relative chromosome length, similarly to total length, was not able to differentiate between S. cereanum (‘Kriszta’) chromosomes. The comparison between the parental rye species revealed statistically significant differences in the short arm length (S) of 1R, 2R, 6R, and 7R, long arm length (L) of 6R, total chromosome length (TL) of 1R and 6R, arm ratio (AR) as well as the centromeric index (CI) of 1R, 2R, 3R, and 7R. The S, L, TL, RL, AR, and CI values of ‘Kriszta’ were not statistically different from those of the parental genotypes except for the AR and CI values of chromosome 3R. The values were either equal to those of one of the parents or were the mean of them (Table 2). The idiogram (Fig. 1) constructed from the data of Table 2 clearly shows that the morphology of the ‘Kriszta’ chromosome set is not identical with either that of Várda or that of R797.
FISH karyotyping
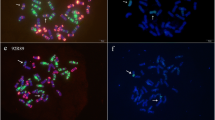
The probe pSc119.2 shows altered in situ hybridization patterns on the ‘Kriszta’ chromosomes relative to those on ‘Várda’ and R797 chromosomes (Fig. 2). One member of the ‘Kriszta’ 1R chromosome pair lacks the terminal signal on the short arm, the subterminal signal on both arms of chromosome 2R and the long arm of 3R is identical to that detected on the long arm of the R797 2R chromosome, and one homolog of the 4R chromosome pair has a terminal signal, which is missing from the chromosome 4R of both ‘Várda’ and R797.
Identification of the short chromosome arms of ‘Kriszta’ using the combination of probes pSc119.2 and (AAC)5 (Fig. 3a) is feasible.
1RS: Being the only rye chromosome carrying a satellite, it can be identified even without FISH signals.
2RS: Although its pSc119.2 pattern is not unique (as mentioned above), it can be unambiguously distinguished from the chromosome arms 3RS, 4RS, 6RS, and 7RS by the multiple (AAC)5 signals, and the 5RS arm also by its length.
3RS and 4RS: Their terminal pSc119.2 signal is similar, but the strong and narrow (AAC)5 signal near the centromere on the 3RS is distinguishable from the broad signal covering half of the chromosome arm 4RS.
5RS: This is the shortest ‘Kriszta’ chromosome arm and it has the strongest terminal pSc119.2 signal. Its (AAC)5 pattern showing a weak band proximal to the centromere and another one in intercalary position is also unique.
6RS: Its two homologs show heteromorphic hybridization patterns. 6RS having a strong pSc119.2 signal in the terminal and another one in the subterminal position is unique, so it can be distinguished from the other short chromosome arms even without the use of an additional FISH probe. 6RS having only a strong terminal signal like that on the 5RS shows a very strong proximal (AAC)5 pattern, which enables us to differentiate between 5 and 6RS.
7RS: Contrary to the other chromosome arms, it has 3 (stronger or weaker) pSc119.2 hybridization sites, thus can be easily identified.
Unambiguous identification of the long arms of ‘Kriszta’ chromosomes with the probes pSc119.2 and (AAC)5 is not possible. There are only two long arms distinguishable from the other ones. 1RL shows a strong terminal pSc119.2 signal, which is the strongest detectable on the long arms, and a weaker one at a distal intercalary position, while 4RL is the only chromosome arm that carries a strong terminal (AAC)5 signal. Although the probe (AAC)5 hybridizes to the 3RL at a subterminal position, the detectability of this faint signal highly depends on the quality of the FISH. The single (AAC)5 band proximal to the centromere is similar on 1RL, 2RL, 5RL, 6RL, and 7RL, and has no diagnostic value when these arms are present e.g. in chromosome translocations. Based on the hybridization sites of the repetitive sequence pSc119.2, the following long arms cannot be differentiated: 2RL and 3RL showing a single subterminal band, 5RL and one of the homologs of 4RL and 6RL having a terminal and an intercalary signal at the same position, as well as 7RL and the other homolog of 6RL (Fig. 2).
Besides (AAC)5, five of the twelve microsatellite sequences generated distinct fluorescence signals on the ‘Kriszta’ chromosomes. Oligonucleotides (ACG)n, (CAC)n, and (CAG)n, apart from the differences in intensity, provided the same pattern as the probe (AAC)5. Oligonucleotides (AGG)n and (GAA)n showed a specific signal on the long arm of chromosome 5R at the terminal position, but only on one member of the chromosome pair. Probe (GAA)n also showed a single intercalary band on one of the homologs of 4RL (Fig. 3b). Probes (AC)n, (AG)n, (ACT)n, (ATT)n, (CAT)n, and (GCC)n did not hybridize to the ‘Kriszta’ chromosomes.
FISH with the repetitive sequence pSc200 located strong terminal hybridization signals on both arms of chromosomes 1R, 2R, and 3R, on the short arm of 7R, and one of the homologs of 5RS, but did not show distinct signals on the chromosomes 4R, 6R, and one member of the chromosome arm pair 5RS. A specific subterminal hybridization site on the chromosome arm 5RL and a specific weak terminal signal on the chromosome arm 7RL were also observable (Fig. 3a).
The probe pSc250 showed a terminal FISH band on the chromosome arms 1RS, 4RS, 5RS, 6RS, and one of the 3RS homologs, together with a weak one on 1RL and a strong one on 3RL. On the other chromosome arms, there were no detectable hybridization signals.
Discussion
As early as 1960, it was pointed out that a critical identification of the rye chromosomes in addition and substitution lines is an absolute necessity because the presence of individual chromosomes is not always reflected in phenotypic characters (Bhattacharyya and Jenkins 1960). Before the appearance of the Giemsa (C and G) chromosome banding [for review see Schubert et al. (1984)] and fluorescence-based in situ hybridization techniques in plants (Leitch et al. 1991; Schwarzacher et al. 1989), the only way to characterize the rye chromosome complement was the karyotype analysis. After performing studies involving several Secale species, it has been concluded that rye has a symmetrical, 1A-type (Paszko 2006; Stebbins 1971) karyotype composed of only metacentric and submetacentric chromosomes of similar size (Cuadrado and Jouve 2002; Masoud and Ali-Jarrahei 2008). Our measurements confirmed this conclusion. Both ‘Kriszta’ and the parental rye genotypes contain one satellite (1R), 3 metacentric (2R, 3R, and 7R), and 3 submetacentric (4R, 5R, and 6R) chromosomes with arm ratios and centromeric indices correlating with the nomenclature described by Levan et al. (1964), even though the arm ratios of the metacentric chromosomes of the cultivated ‘Várda’ differ significantly from those of the wild R797 genotype. Based on the total length data, the karyotype of S. strictum ssp. anatolicum (R797) is different from that of S. cereale as the former contains significantly shorter 1R and 6R than the latter. The total length data and centromere positions of R797 correlates very well with the rye pseudomolecule sizes determined from the recent chromosome-scale genome assembly (high-quality reference genome sequence) of cereal rye inbred line ‘Lo7’ (Rabanus-Wallace et al. 2021, Supplementary note Fig. 4.2.). According to two data set, 1R is the shortest chromosome (~ 720 Mb) followed by 5R (~ 880 Mb), 6R (~ 890 Mb), 7R (~ 900 Mb), 4R (~ 910 Mb), 2R (~ 950 Mb), and 3R (~ 90 Mb). Interestingly, the data concerning ‘Várda’ (S. cereale) do not follow this tendency. The authors speculate that this may be the consequence of changes in the chromosome set caused by selection pressures during the breeding of this cultivar.
The S. cereanum (‘Kriszta’) chromosomes do not differ from those of S. cereale in this parameter except for 6R, which has the same size as 6R of S. strictum ssp. anatolicum. At the same time, ‘Kriszta’ has altered centromere positions, which is reflected in the values of AR and CI. In the present study, the preparation of metaphase plates and the chromosome measurements using digital images was carried out in the same way, thus the differences in the measured parameters can be considered real, not the consequence of varying experimental conditions (Heneen 1962; Sybenga 1959). The results presented above make us conclude that rearrangements may have occurred between (and maybe within) the parental rye chromosomes during the hybridization or the breeding of ‘Kriszta’, consequently, its chromosome complement is not a simple combination of the chromosomes of S. cereale and S. strictum ssp. anatolicum. Furthermore, it cannot be determined by size which chromosome arm is involved in a given translocation or whether it is of ‘Várda’ or wild rye origin.
According to the FISH hybridization patterns of the repetitive DNA sequence pSc119.2, changes also have occurred in the heterochromatin of ‘Kriszta’ chromosomes compared to those of ‘Várda’ and R797 leading to the fact that the chromosome arms of ‘Kriszta’ are even more difficult to identify with this probe. Nevertheless, thanks to the uniqueness of the ‘Kriszta’ chromosomes, some DNA sequences have proved to be suitable to distinguish between chromosome arms. Studies on rye chromosome variability in wheat-rye addition and substitution lines (Alkhimova et al. 1999) and on detection alterations in cultivated rye chromosomes in newly synthesized wheat-rye allopolyploids (Fu et al. 2010) showed that pSc200 FISH hybridization sites were located at both telomeres or subtelomeres of all 14 chromosomes of S. cereale. Investigating evolutionary trends of different repetitive DNA sequences, Cuadrado and Jouve (2002) found that among the subspecies of S. strictum only anatolicum exhibited pSc200 signals on both arms of each chromosome. Contrary to these observations, hybridization sites are absent on the 4R and 6R chromosomes of ‘Kriszta’. The 5RL homologs show the presence of hybridization at the subtelomeric position, which is different from the telomeric signals on the chromosome arms 1RL, 2RL, 3RL, and 7RL. Consequently, the hybridization pattern of 5RL is a prominent landmark that enables the unambiguous identification of this ‘Kriszta’ chromosome long arm. Taking into consideration that the simple sequence repeat oligonucleotide (AAC)5 hybridizes specifically to 4RL at a telomeric position in ‘Kriszta’, as it was described also in the case of different Secale species and subspecies (Cuadrado and Jouve 2002), the DNA probe combination pSc200-(AAC)5 is suitable to differentiate between chromosome arms 4RL, 5RL, 6RL, and 7RL of ‘Kriszta’.
The distribution of the tandem repeat pSc250 on ‘Kriszta’ chromosomes is in line with the previous observations in S. cereale (Alkhimova et al. 1999) and its wild relatives (Cuadrado and Jouve 2002), namely that the strongest hybridization can be detected at the telomeric position of the short arms of all chromosomes and the long arms of chromosomes 1R and 3R. In ‘Kriszta’, 1RL shows a faint, while 3RL a strong (double) signal. Based on this, the chromosome arms 1RL, 2RL (no signal), and 3RL are also distinguishable. Unfortunately, the oligonucleotide sequences, except for (AAC)5, cannot be used for identifying individual ‘Kriszta’ chromosome long arms. Although the telomeric (AGG)n signal on 5RL, and the interstitial and telomeric (GAA)n signals on 4RL and 5RL, respectively, are of diagnostic value (differentiating them from the other ‘Kriszta’ chromosomes), they can be detected in only one of the homologs. This heterozygosity for the distribution of microsatellite motifs has been described in most (the allogamous) rye species and subspecies (Cuadrado and Jouve 2002).
The loss of the rye-specific pSc200 FISH signals in the ‘Kriszta’ 4R and 6R chromosomes is an interesting phenomenon. The massive blocks of subtelomeric heterochromatin of rye (Secale spp.) chromosomes have originated from the combined activity of transposable elements and the expansion of tandemly repeated DNA sequences (Evtushenko et al. 2016). The subtelomeric tandem repeats, including pSc200, are considered to be fast-evolving components of the eukaryotic genomes (Vershinin et al. 1996). There are pieces of evidence that the copy number of these repeats can change rapidly not only on an evolutionary scale. A study on the alteration of the subtelomeric heterochromatin in newly produced wheat-Aegilops allopolyploids (Han et al. 2005) showed drastic loss of hybridization signals of the tandem repetitive DNA sequence pGc1R-1 by the second selfed generation. Similar results were published by Tang et al. (2008) and Fu et al. (2010) when investigating newly synthesized wheat-rye F1 hybrids and amphidiploids using the rye-specific pSc200. It was speculated by them that the role of the elimination of parental DNA repeats upon allopolyploidy is to eliminate or mitigate the genomic incompatibility of parental genomes (Han et al. 2005). Nevertheless, ‘Kriszta’ is the progeny of a hybrid between two Secale species carrying (almost) the same R genome, thus, there were probably no compatibility problems during the hybridization. Consequently, the reason for the decrease in the content of the subtelomeric heterochromatin (indicated by the absence of pSc200 signals) is not clear.
The results of the present study indicate, that an adequate pair-wise combination of the DNA sequences pSc119.2, pSc200, pSc250, and (AAC)5 makes it possible to identify any of the long arms of S. cereanum chromosomes in wheat- ‘Kriszta’ translocations. The translocated chromosomes with specific FISH hybridization patterns later can be sorted by flow cytometry (Kubaláková et al. 2003) based on FISHIS (FISH in suspension) (Giorgi et al. 2013) and used in chromosome-based genomics (Doležel et al. 2007). This makes it possible in the future to sequence chromosome(arm)-specific DNAs and isolate genes originated from the exotic genome of S. cereanum.
Availability of data and material
Not applicable.
Code availability
Not applicable.
References
Achrem M, Kalinka A, Rogalska SM (2014) Assessment of genetic relationships among Secale taxa by using ISSR and IRAP markers and the chromosomal distribution of the AAC microsatellite sequence. Turk J Bot 38:213–225. https://doi.org/10.3906/bot-1207-26
Alkhimova AG, Heslop-Harrison JS, Shchapova AI, Vershinin AV (1999) Rye chromosome variability in wheat–rye addition and substitution lines. Chromosome Res 7:205–212. https://doi.org/10.1023/A:1009299300018
Bedbrook JR, Jones J, O’Dell M, Thompson RD, Flavell RB (1980) A molecular description of telomeric heterochromatin in Secale species. Cell 19:545–560. https://doi.org/10.1016/0092-8674(80)90529-2
Bhattacharyya NK, Jenkins BC (1960) Karyotype analysis and chromosome designations for Secale cereale L. “Dakold.” Can J Genet Cytol 2:268–277. https://doi.org/10.1139/g60-027
Contento A, Heslop-Harrison JS, Schwarzacher T (2005) Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet Genome Res 109:34–42. https://doi.org/10.1159/000082379
Cuadrado A, Jouve N (2002) Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. J Hered 93:339–345. https://doi.org/10.1093/jhered/93.5.339
Cuadrado A, Schwarzacher T (1998) The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107:587–594. https://doi.org/10.1007/s004120050345
Doležel J, Kubaláková M, Paux E, Bartos J, Feuillet C (2007) Chromosome-based genomics in the cereals. Chromosome Res 15:51–66. https://doi.org/10.1007/s10577-006-1106-x
Endo TR, Gill BS (1984) Somatic karyotype, heterochromatin distribution, and nature of chromosome differentiation in common wheat Triticum aestivum L em Thell. Chromosoma 89:361–369. https://doi.org/10.1007/BF00331253
Evtushenko EV, Levitsky VG, Elisafenko EA, Gunbin KV, Belousov AI, Šafář J, Doležel J, Vershinin AV (2016) The expansion of heterochromatin blocks in rye reflects the co-amplification of tandem repeats and adjacent transposable elements. BMC Genomics 17:337. https://doi.org/10.1186/s12864-016-2667-5
Fu S, Tang Z, Ren Z (2010) Inter- and intra-genomic transfer of small chromosomal segments in wheat-rye allopolyploids. J Plant Res 123:97–103. https://doi.org/10.1007/s10265-009-0264-2
Füle L, Hódos-Kotvics G, Galli Z, Ács E, Heszky L (2005) Grain quality and baking value of perennial rye (cv. ‘Perenne’) of interspecific origin (Secale cereale × S. montanum). Cereal Res Commun 33:809–816. https://doi.org/10.1556/CRC.33.2005.2-3.152
Giorgi D, Farina A, Grosso V, Gennaro A, Ceoloni C, Lucretti S (2013) FISHIS: fluorescence in situ hybridization in suspension and chromosome flow sorting made easy. PLoS ONE 8:e57994. https://doi.org/10.1371/journal.pone.0057994
Han F, Fedak G, Guo W, Liu B (2005) Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 170:1239–1245. https://doi.org/10.1534/genetics.104.039263
Heneen W (1962) Chromosome morphology in inbred rye. Hereditas 48:182–200. https://doi.org/10.1111/j.1601-5223.1962.tb01806.x
Kirov I, Khrustaleva L, van Laere K, Soloviev A, Sofie M, Romanov D, Fesenko I (2017) DRAWID: user-friendly java software for chromosome measurements and idiogram drawing. Comp Cytogenet 11:747–757. https://doi.org/10.3897/CompCytogen.v11i4.20830
Kotvics G (1970) Investigations on increasing the protein content of Secale cereale L. In: Bálint A (ed) Protein growth by plant breeding. Akadémiai Kiadó, Budapest, Hungary, pp 89–90
Kruppa J (2001) Rozs és triticale nemesítés és tájtermesztés eredményei. PhD, Universitiy of Debrecen
Kubaláková M, Valárik M, Barto J, Vrána J, Cíhalíková J, Molnár-Láng M, Doležel J (2003) Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46:893–905. https://doi.org/10.1139/g03-054
Leitch IJ, Leitch AR, Heslop-Harrison JS (1991) Physical mapping of plant DNA sequences by simultaneous in situ hybridization of two differently labelled fluorescent probes. Genome 34:329–333. https://doi.org/10.1139/g91-054
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Masoud S, Ali-Jarrahei S (2008) Cytogenetical studies of some species of the genus Secale L. (Poaceae) in Iran. Caryologia 61:182–189. https://doi.org/10.1080/00087114.2008.10589628
McIntyre CL, Pereira S, Moran LB, Appels R (1990) New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33:635–640. https://doi.org/10.1139/g90-094
Molnár-Láng M, Linc G, Sutka J (1996) Transfer of the recessive crossability allele kr1 from Chinese Spring into the winter wheat variety Martonvásári 9. Euphytica 90:301–305. https://doi.org/10.1007/BF00027480
Nagaki K, Tsujimoto H, Isono K, Sasakuma T (1995) Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome 38:479–486. https://doi.org/10.1139/g95-063
Paszko B (2006) A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol 258:39–48. https://doi.org/10.1007/s00606-005-0389-2
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593. https://doi.org/10.1139/g97-077
Rabanus-Wallace MT, Hackauf B, Mascher M, Lux T, Wicker T, Gundlach H et al (2021) Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat Genet 53:564–573. https://doi.org/10.1038/s41588-021-00807-0
Said M, Hřibová E, Danilova TV, Karafiátová M, Čížková J, Friebe B, Doležel J, Gill BS, Vrána J (2018) The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor Appl Genet 131:2213–2227. https://doi.org/10.1007/s00122-018-3148-9
Schneider A, Linc G, Molnár-Láng M (2003) Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breeding 122:396–400. https://doi.org/10.1046/j.1439-0523.2003.00891.x
Schneider A, Rakszegi M, Molnár-Láng M, Szakács É (2016) Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rust resistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theor Appl Genet 129:1045–1059. https://doi.org/10.1007/s00122-016-2682-6
Schubert I, Rieger R, Döbel P (1984) G and/or C-bands in plant chromosomes? J Cell Sci 71:111–120. https://doi.org/10.1242/jcs.71.1.111
Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS (1989) In situ localization of parental genomes in a wide hybrid. Ann Bot 64:315–324. https://doi.org/10.1093/oxfordjournals.aob.a087847
Stebbins GL (1971) Chromosomal evolution in higher plants. Contemporary biology. Edward Arnold, London
Sybenga J (1959) Some sources of error in the determination of chromosome length. Chromosoma 10:355–364. https://doi.org/10.1007/BF00396578
Szakács É, Szőke-Pázsi K, Kalapos B, Schneider A, Ivanizs L, Rakszegi M, Vida G, Molnár I, Molnár-Láng M (2020) 1RS arm of Secale cereanum “Kriszta” confers resistance to stripe rust, improved yield components and high arabinoxylan content in wheat. Sci Rep 10:1792. https://doi.org/10.1038/s41598-020-58419-3
Tang Z-X, Fu S-L, Ren Z-L, Zhou J-P, Yan B-J, Zhang H-Q (2008) Variations of tandem repeat, regulatory element, and promoter regions revealed by wheat-rye amphiploids. Genome 51:399–408. https://doi.org/10.1139/G08-027
Vershinin AV, Schwarzacher T, Heslop-Harrison JS (1995) The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell 7:1823–1833. https://doi.org/10.1105/tpc.7.11.1823
Vershinin AV, Alkhimova EG, Heslop-Harrison JS (1996) Molecular diversification of tandemly organized DNA sequences and heterochromatic chromosome regions in some Triticeae species. Chromosome Res 4:517–525. https://doi.org/10.1007/BF02261779
Zhou J, Yang Z, Li G, Liu C, Tang Z, Zhang Y, Ren Z (2010) Diversified chromosomal distribution of tandemly repeated sequences revealed evolutionary trends in Secale (Poaceae). Plant Syst Evol 287:49–56. https://doi.org/10.1007/s00606-010-0288-z
Acknowledgements
This research was funded by the National Research, Development and Innovation Office—NKFIH, K 119387
Funding
Open access funding provided by ELKH Centre for Agricultural Research. National Research, Development and Innovation Office—NKFIH, K 119387.
Author information
Authors and Affiliations
Contributions
ÉS conceived the study, KS-P performed the cytomolecular studies, ET carried out the chromosome measurements, KS-P and ÉS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Szőke-Pázsi, K., Türkösi, E. & Szakács, É. Chromosome morphology and cytomolecular characteristics of the perennial rye cultivar ‘Kriszta’. CEREAL RESEARCH COMMUNICATIONS 50, 789–796 (2022). https://doi.org/10.1007/s42976-021-00233-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-021-00233-2