Abstract
Apple scab resistance genes, HcrVf1 and HcrVf2, were isolated including their native promoter, coding and terminator sequences. Two fragment lengths (short and long) of the native gene promoters and the strong apple rubisco gene promoter (PMdRbc) were used for both HcrVf genes to test their effect on expression and phenotype. The scab susceptible cultivar ‘Gala’ was used for plant transformations and after selection of transformants, they were micrografted onto apple seedling rootstocks for scab disease tests. Apple transformants were also tested for HcrVf expression by quantitative RT-PCR (qRT-PCR). For HcrVf1 the long native promoter gave significantly higher expression that the short one; in case of HcrVf2 the difference between the two was not significant. The apple rubisco gene promoter proved to give the highest expression of both HcrVf1 and HcrVf2. The top four expanding leaves were used initially for inoculation with monoconidial isolate EU-B05 which belongs to race 1 of V. inaequalis. Later six other V. inaequalis isolates were used to study the resistance spectra of the individual HcrVf genes. The scab disease assays showed that HcrVf1 did not give resistance against any of the isolates tested regardless of the expression level. The HcrVf2 gene appeared to be the only functional gene for resistance against Vf avirulent isolates of V. inaequalis. HcrVf2 did not provide any resistance to Vf virulent strains, even not in case of overexpression. In conclusion, transformants carrying the apple-derived HcrVf2 gene in a cisgenic as well as in an intragenic configuration were able to reach scab resistance levels comparable to the Vf resistant control cultivar obtained by classical breeding, cv. ‘Santana’.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most of the present day apple (Malus × domestica) cultivars are susceptible to apple scab which is caused by the fungus Venturia inaequalis. Fruit growers spray on average 15 times in one season to control the disease (Patocchi et al. 2004).
Most of the conventionally bred resistant cultivars can give resistance against race 1 to race 5 (Szankowski et al. 2009) and against race 8 of V. inaequalis. The first report of breaking down of Vf resistance by a new race of V. inaequalis was made by Parisi et al. (1993) and they called this race 6. Later, again an isolate was identified which had overcome the resistance of Vf which was subsequently named as race 7 (Parisi et al. 2004).
Several apple scab resistance genes such as HcrVf (Homologues of C ladosporium fulvum resistance genes of Vf region) (Patocchi et al. 1999), Vr2 (Patocchi et al. 2004), Vd3 (Soriano et al. 2009), Vb (Erdin et al. 2006), Va (Hemmat et al. 2003), Vbj (Gygax et al. 2004) have been identified and were mapped on different linkage groups of the apple genome. However, only the Vf locus has been positionally cloned (Vinatzer et al. 2001) and proved to consists of a gene cluster with four paralogs, namely HcrVf1, HcrVf2, HcrVf3 and HcrVf4 (Xu and Korban 2002). Initially, only the HcrVf2 gene was tested by overexpression using the CaMV35S promoter and nos terminator, in scab susceptible apple plants (Belfanti et al. 2004) and the transgenic apple plants overexpressing HcrVf2 proved to be resistant to apple scab.
Later, Malnoy et al. (2008) transformed Vfa1 (which is the same as HcrVf1) and Vfa2 (which is the same as HcrVf2) with their native promoters and terminators, to susceptible cultivars ‘Galaxy’ and ‘McIntosh’ and showed partial resistance of HcrVf1 and HcrVf2 to a mixture of isolates (Race 1–Race 5) of V. inaequalis. They used a 2 kb promoter for both Vfa1 and Vfa2 and they observed a reduction in sporulation by 50 and 38% in plants with good expression for Vfa1 and Vfa2 respectively. Szankowski et al. (2009) showed complete resistance by inserting only HcrVf2 under the regulation of different lengths of native promoters and the nos terminator into susceptible cv. ‘Gala’. They used also a mixture of V. inaequalis isolates for scab inoculation. This conflict in results prompted us to investigate this in more detail.
Ribulose-1, 5-bisphosphate carboxylase/oxygenase (rubisco) is an abundant protein in plants (Ellis 1979) and rubisco protein contents were correlated with total rubisco small subunit (RbcS) mRNA levels in rice (Suzuki et al. 2009). Using the promoter and terminator of rubisco small subunit gene (rbcS1) of chrysanthemum, expression levels up to sevenfold to eightfold those provided by the constitutively expressed CaMV35S promoter were obtained (Outchkourov et al. 2003). The combination of the apple rubisco promoter (PMdRbc) and terminator (TMdRbc) with the gus reporter gene resulted in expression levels similar to the one provided by the CaMV35S promoter in combination with the nos terminator (Schaart et al. 2011).
Transgenes, consisting of foreign genes or parts of foreign genes, are frequently used in GM plants and are a new gene pool for plant breeding. Nowadays, cisgenes defined as genes from the crop plant itself or from crossable species, with their native regulatory elements including introns in normal sense orientation (Schouten et al. 2006a) receive increasing interest. Cisgenic plants do not contain foreign genes by definition. Cisgenes are already present in the species or in cross-compatible relatives and therefore cisgenesis does not alter the gene pool used in breeding a crop (Schouten et al. 2006b). Intragenes are composed of different genetic elements originating from the crop species itself or from crossable plant species (Rommens et al. 2007). Genetic elements include promoters, coding sequence, terminators and DNA sequences that are similar to T-DNA borders from Agrobacterium tumefaciens (Jacobsen and Schouten 2008).
In this study we report on the role and resistance spectrum of the cisgenes HcrVf1 and HcrVf2 together with their native promoters in two lengths (short and long) and their native terminator in conferring resistance to apple scab. As a reference, the two HcrVf genes were combined with regulatory sequences of the apple rubisco gene and are therefore referred to as intragenes. The correlations between inserted gene copy number and gene expression and between gene expression and fungal sporulation were investigated. To our knowledge this is first detailed study of expression profiling of two individual Vf resistance genes using different isolates of V. inaequalis identifying their resistance level and spectrum.
Materials and methods
Gene amplification
Preparation of HcrVf constructs with native promoters
An apple BAC library was constructed from the genomic DNA of the breeding line 1980-015-025 which harbors several scab resistance genes such as HcrVf, V25, Vd3 (Soriano et al. 2009). The BAC library construction was carried out as previously described by Rouppe van der Voort et al. (1999). Primers described by Xu and Korban (2002) (Table 1) were used to identify individual BAC clones from BAC pools positive for HcrVf1 and HcrVf2. One BAC clone containing HcrVf1 and one containing HcrVf2 were sequenced at GATC Biotech AG (Konstanz, Germany). Sequenced contigs were aligned with the published HcrVf1 (Gene bank accession number AY397723) and HcrVf2 (Gene bank accession number AJ297740) sequences using the software SeqMan in DNASTAR® version 7.0. On the basis of the recommendations by Silfverberg-Dilworth et al. (2005) sequences up to 312 and 288 bp upstream of the transcription start site were used as short promoters (SP) for HcrVf1 and HcrVf2, respectively. Similarly, sequences up to 480 and 437 bp downstream of the stop codon were used as terminators for HcrVf1 and HcrVf2, respectively. Sequences of 1,990 and 2,000 bp upstream of the transcription start site were used as long promoters (LP) for HcrVf1 and HcrVf2, respectively, and terminators were the same as in case of SPHcrVf1 and SPHcrVf2. The details of gene sequences are shown in Fig. 1.
Constructs used for plant transformation. P Promoter, cds coding sequence, T terminator. The numbers in parentheses indicate the lengths of promoter, coding sequence and terminator in basepairs. Vertical stripes represent apple rubisco promoter and horizontal stripes represent apple rubisco terminator. All LP and SP constructs represent stretches cloned as a whole; the PMdRbc constructs represent new combinations
The fragments were amplified using Phusion® DNA polymerase (Finnzymes, Espoo, Finland) and BAC DNA as template. The primers used were extended with AscI and MluI restriction site sequences for HcrVf1 at 5′ and 3′ ends respectively, and AscI and PacI restriction site sequences for HcrVf2 at 5′ and 3′ ends respectively. The primers are listed in Table 1. The PCR reaction included 5× Phusion® HF buffer, 5 mM dNTPs, 10 μM forward and reverse primers (each), 0.2 U Phusion® DNA polymerase making a total volume of 20 μl. The PCR conditions were as follows: 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 56°C for 30 s and 72°C for 4 min and final extension at 72°C for 10 min to generate whole gene amplification of HcrVf1 and HcrVf2. The PCR fragments were cloned in pGEMT-Easy (Promega, WI, USA) and the cloned genes were sequenced to confirm that the complete gene sequences were free of PCR-errors. Subsequently, these fragments were subcloned into the binary, marker-free vector pMF1 (Schaart et al. 2010) in the multiple cloning site using AscI and PacI restriction sites.
Preparation of HcrVf constructs with apple rubisco gene regulatory elements
PMdRbc and TMdRbc sequences were amplified using sequence specific primers (Schaart et al. 2011). Sequence amplification was done using primers with restriction site sequences of PacI and XmaI at their 5′ and 3′ ends respectively for PMdRbc and KpnI and AscI at 5′ and 3′ ends respectively for TMdRbc. PMdRbc and TMdRbc sequences were cloned in pGEMT-Easy and subsequently combined so that they were separated by XmaI and KpnI and flanked by PacI and AscI. HcrVf1 and HcrVf2 genes were amplified using gene specific primers (Table 1) giving restriction sites XmaI at the 5′ end and KpnI at 3′ end and cloned in a vector. These gene fragments were cloned in pGEMT-Easy and confirmed by sequencing to make sure that the sequences are PCR-error-free. Both HcrVf sequences were excised using XmaI and KpnI, and subsequently subcloned into the PMdRbc and TMdRbc construct. Then the PMdRbc–HcrVf–TMdRbc fragments were subcloned as PacI-AscI fragment into the binary vector pMF1 (Schaart et al. 2010). The gene constructs used in plant transformation are given in Fig. 1.
Plant transformation and regeneration
The resulting pMF1 with HcrVf genes under control of native or rubisco gene regulatory elements were transformed to the supervirulent Agrobacterium tumefaciens strain AGL0 (Lazo et al. 1991). Transformation of apple was basically performed as described by Puite and Schaart (1996) with some minor modifications. The top four leaves of 4-week-old, in vitro propagated ‘Gala’ shoots were used as explants and transferred to a sterile Petri dish filled with 15 ml A. tumefaciens suspension for inoculation. The leaves were cut into 0.5 cm2 pieces and incubated for 30 min. Then, the explants were dried on a sterile filter paper and transferred for co-cultivation to shoot induction medium (SIM) consisting of MS medium with vitamins, micro and macro elements (Murashige and Skoog 1962), 3% (w/v) sorbitol, 9.9 μM thidiazuron (TDZ), 0.5 μM NAA, 13.3 μM BAP, 0.3% (w/v) Gelrite, pH 5.8 for 4 days. After 4 days of cocultivation the explants were transferred to selective regeneration medium, i.e. SIM with 100 mg/l kanamycin for selection of transformants and 250 mg/l cefotaxim to eliminate A. tumefaciens. Then, the leaf explants were cultured in the dark at 24°C and subcultured at 4-weekly intervals on fresh selective regeneration medium. After culturing in the dark for 12 weeks, when callus had been produced on the explants and shoot-like structures started emerging from the calli, the explants were exposed to diffused light and later gradually to full light conditions of 7,000 lux (16 h/day) to avoid direct light stress (Espley et al. 2007). When the shoots were big enough, they were isolated and transferred to shoot elongation medium (SEM) containing MS with vitamins, micro and macro elements, 1% (w/v) galactose, 2% (w/v) sucrose, 0.5 μM NAA, 4.4 μM BAP, 0.9% (w/v) Daishin agar, pH 5.8 for 4 weeks with 16 h light/8 h dark at 24°C; normal developing shoots, i.e. putative transformants, were subsequently multiplied on shoot propagation medium (SPM) containing MS with vitamins, micro and macro elements, 3% (w/v) sucrose, 96 mg/l FeEDDHA, 3.1 μM BAP, 0.9% (w/v/) Daishin agar, pH 5.8.
Molecular analysis of transformants
Genomic DNA from putative transformants was isolated as described by Jaccoud et al. (2001). All the putative transformants were analyzed for the presence of inserted genes using HcrVf1 and HcrVf2 gene specific primers and with nptIII, and trfA primers for the presence of pMF1 binary vector backbone (primers mentioned in Table 1). The PCR reaction included 10× Supertaq™ buffer, 5 mM dNTPs, 10 μM each of forward and reverse primers, 0.5 U Supertaq™ DNA polymerase making a total volume of 20 μl. The PCR conditions were as follows: 96°C for 5 min, followed by 30 cycles of 96°C for 30 s, 55°C for 45 s and 72°C for 1 min and 30 s and final extension at 72°C for 10 min to generate an internal fragment of 505 bp for both HcrVf1 and HcrVf2. Similar PCR conditions were used for nptIII and trfA to generate a fragment of 475 and 345 bp, respectively.
T-DNA integration was analyzed by Southern hybridization, as described by Southern (1975). For this, 20 μg of genomic DNA was digested overnight at 37°C with the restriction enzyme BglII, separated on a 1% (w/v) agarose gel, and finally blotted onto a positively charged Hybond N+ nylon membrane (Amersham, little Chalfont, UK). The nptII probe was prepared using primers (mentioned in the Table 1) amplifying a fragment of 721 bp. The nptII probe was labeled with 32P dCTP (Amersham, Little Chalfont, UK) using Rediprime II random prime labelling system (Amersham) and used for hybridization. Untransformed cv. ‘Gala’ was used as negative control and a plasmid containing the nptII gene was used as positive control.
Micrografting
For each construct, six independent transformation events (unless stated otherwise) were selected and multiplied in vitro to provide six replicates per event for HcfVf gene expression and scab resistance studies. Apple seedlings derived from a cross between Golden Delicious × Baskatong or from a cross between Elstar × Baskatong were obtained from the NAKTuinbouw (Horst, The Netherlands) and were used as rootstock in micrografting experiments. The ‘in vitro’ grown plants were grafted directly onto these rootstocks as described by Lane et al. (2003) and grown in the greenhouse. Four to six weeks after micrografting, the grafted plants had developed at least four, new, young leaves and were ready for scab inoculations. At this stage non-infected plant material was harvested for DNA and RNA extraction.
RNA isolation and quantitative RT-PCR
Total RNA was isolated from young leaves of the micrografted plants using the RNeasy mini kit (Qiagen, Carlsbad, CA, USA). From all replicates per event, leaves were harvested and pooled to get sufficient amounts of plant material for RNA isolation and for further quantification; hence, each RNA sample constituted the average of the six biological repeats for each event. The RNA samples were run on a 1% (w/v) agarose gel to determine the RNA quality. An aliquot total RNA of 2 μg was used to treat with DNaseI (Invitrogen Carlsbad, CA, USA) and subsequently reverse transcribed with a blend of oligo (dT) and random primers to synthesis cDNA using iScript first strand cDNA synthesis kit (Bio-rad Hercules, CA, USA). The reactions were performed according to the manufactures guidelines. Quantitative RT-PCR (qRT-PCR) was carried out to check the expression levels of the HcrVf genes and to correlate the expression with the gene copy number and also to correlate inserted gene expression with fungal sporulation. Cv. ‘Santana’ is a scab resistant variety carrying both HcrVf1 and HcrVf2 genes introduced by conventional breeding and the expression of HcrVf1 and HcrVf2 in transgenic cv. Gala plants was measured as fold change by comparing with expression in ‘Santana’. Untransformed ‘Gala’ does not harbour functional HcrVf scab resistance genes.
The qRT-PCR was performed with iQ SYBR® green super mix (Bio-rad) with MyiQ Single Color Real time detection system. As endogenous reference the β-actin gene (Accession number DT002474) was used. Each extract was checked twice (technical repeats). The primer sequences used for qRT-PCR are presented in Table 1. Primer efficiencies were calculated through different dilution series of cDNA (Rebrikov and Trofimov 2006). All the PCR reactions were carried out in duplicates. HcrVf gene expression levels were analyzed by using relative quantification method i.e. 2−ΔΔCt method through qRT-PCR (Li et al. 2004). The threshold cycle (Ct) is the PCR cycle at which a statistically significant increase in the transcript is first detected.
The normalized Ct value difference (ΔCt) was calculated from Ct (HcrVf)-Ct (β-actin) for all the reactions. The ΔCt value of ‘Santana’ which has one copy of the natural HcrVf genes was chosen as reference sample and for all samples relative ΔΔCt values were calculated using ΔCt (reference sample ‘Santana’) − ΔCt (target sample). Finally the transgene expression levels were determined as fold change using the formula 2−ΔΔCt.
Scab resistance evaluation against EU-B05
Scab disease tests were conducted in a temperature and humidity controlled greenhouse. A conidial suspension of V. inaequalis isolate EU-B05 (Bus et al. 2005) containing 5 × 105 conidia/ml was prepared. During the inoculations, the suspensions were checked for germination in vitro and this was generally found to be more than 90%. The top four young, expanding leaves of the scions were used for inoculation with the well-characterized, monoconidial isolate EU-B05. For each transgenic event six replicates (one plant per replicate) were used unless stated otherwise. Inoculated plants were kept in a plastic tunnel in the dark for 48 h at 20°C and 100% relative humidity. After this period, the plants were transferred outside the tunnel, but still kept in the same greenhouse compartment with the temperature set at 19°C during day and 16°C during night, day length of 16 h and relative humidity of 85%.
Disease symptoms were assessed macroscopically 17 days post inoculation and classified in eight classes as reported by Durel et al. (2003), indicative for the amount of sporulation as follows: class 0, 0% of sporulation; class 1, 1–2% sporulation; class 2, 2–5% sporulation; class 3, 5–10% sporulation; class 4, 10–25% sporulation; class 5, 25–50% sporulation; class 6, 50–75% sporulation; class 7, 75–100% sporulation. This scale was adapted from Croxall et al. (1952).
Scab resistance evaluation against different isolates
Based on the results of scab resistance evaluation against the EU-B05 scab experiment, plant transformants were selected from all the representatives of the gene constructs to check the resistance spectrum of the HcrVf genes. Six isolates, of which EU-B05, 1639, US-3 and NZ 188 (Bus et al. 2005) are avirulent isolates and EU-D42 (Bus et al. 2005) and EU-NL05 (Parisi et al. 2004) are virulent isolates of V. inaequalis for cv. ‘Santana’ and other Vf based resistant varieties, were used to study the resistance spectrum.
Both quantitative (as described above) and qualitative (Chevalier et al. 1991; Szankowski et al. 2009) scales were used for scoring 21 days after inoculation.
Statistical analysis
For statistical analysis of the relative expression, each event was taken as experimental unit with the leaves of the six replicates (biological repeats) pooled before RNA isolation. Here, the experimental design was unbalanced due to variation in number of replications. The data were analyzed through Student’s t test (unpaired) by comparing two promoters at a time.
For the statistical analysis of scab resistance, each plant was taken as experimental unit. The scab resistance evaluation against EU-B05 was conducted with Randomized Complete Block Design (RCBD). Six replicates were used for each transformation event; for each construct 3–6 independent lines were taken. In total, 31 events and 198 plants were investigated. All the statistical analyses were performed using Genstat® 12 (Genstat® 2009). Since the treatments were not balanced, an ANOVA could not be used. In stead we used the linear mixed model (LMM) procedure. The expression data were log transformed and correlation was studied between phenotypic data and log transformed expression data using Spearman’s correlation.
The scab resistance evaluation against different isolates was also performed as a Randomized Complete Block Design (RCBD). The genotypes and isolates were used as the experimental units. The number of replicates used in this experiment was four and in total 201 plants were used in this analysis. In some genotypes there were not equal number of plants per replication and there were not equal number of plants for inoculation with some isolates. Therefore the experimental setup became unbalanced, so a linear mixed model (LMM) was fitted and used for analysis.
Results
Production of transformation vectors carrying HcrVf genes
A BAC library of the genotype 1980-015-025, containing the Vf resistance, was screened for clones, harboring HcrVf1 and HcrVf2. Four independent BAC pools namely #12 and #199 for HcrVf1, #105 and #228 for HcrVf2 were identified by PCR and the BAC clones harboring these genes were isolated from the pools. The complete gene sequences including promoters of two different lengths, coding sequences and terminators as single stretches for both genes were amplified (Figs. 1, 2) using the BAC clone DNA as template and equipped with the appropriate restriction sites for further cloning. The fragments with the expected sizes (boxed fragments in Fig. 2) were isolated for further processing. Short promoter (SP) HcrVf1, long promoter (LP) HcrVf1, short promoter (SP) HcrVf2 and long promoter (LP) HcrVf2 were then cloned into the transformation vector for production of marker-free plants, pMF1 (Schaart et al. 2004). In addition to this, the coding regions of both HcrVf genes were combined in two constructs with the apple rubisco gene promoter (PMdRbc) and terminator (TMdRbc) and introduced into pMF1 as AscI-PacI fragments. The individual binary vectors were introduced into Agrobacterium strain AGL0 and used for plant transformation.
Production and molecular analysis of transformants
Six independent transgenic lines were selected for SPHcrVf1, LPHcrVf1, PMdRbc HcrVf1 and PMdRbc HcrVf2 and four and three independent transgenic lines were selected for SPHcrVf2 and LPHcrVf2, respectively. All the HcrVf1 and HcrVf2 plant transformants were checked for the presence of the respective genes using HcrVf1- and HcrVf2-specific primers and proved to be positive (data not shown). Twenty six out of 31 transformants showed the absence of the pMF1 vector backbone which was checked using the primers for nptIII and trfA (primer details given in Table 1, data not shown). T-DNA integration and copy number determination were analyzed by Southern hybridization (Table 2, Fig. 3). Out of 26 transformants taken for estimation of inserted gene copies, 17 have a single T-DNA insert, seven were having two inserts, while two transformants could not be analyzed.
Quantitative RT-PCR
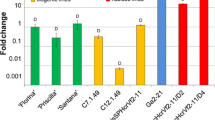
There was a wide variation in the expression of the HcrVf genes, controlled by different promoters. The relative expression of the genes is given in the Table 2. The expression levels of SPHcrVf1, LPHcrVf1 and PMdRbc HcrVf1 were in the range of 0.4–2.3, 5.5–20, and 223–762-fold respectively, compared to HcrVf1 expression in the resistant control cv. ‘Santana’ obtained by conventional breeding. The expression levels of SPHcrVf2, LPHcrVf2 and PMdRbc HcrVf2 were in the range of 0.23–1.2, 1.1–7.1, and 57–163 respectively compared to HcrVf2 expression in ‘Santana’.
Statistical analysis of the data showed that HcrVf1 under control of its LP showed a significantly higher expression than in case of the SP. The apple rubisco promoter gave the highest (355 times HcrVf1 expression in ‘Santana’) expression levels (Fig. 4a). In case of HcrVf2, the performances of SP and LP were not significantly different, although LP generally gave a slightly higher expression. Still, expression by PMdRbc was significantly higher (94 times the HcrVf2 expression in ‘Santana’) than by SP and LP for HcrVf2 (Fig. 4b).
a and b Relative expression of HcrVf genes under control of different gene promoters. SP short native promoter, LP long native promoter, P MdRbc apple rubisco promoter. Santana as reference was set at 1 (see also Table 2). For visual convenience two scales have been plotted together. Two oblique lines indicate change in the scale in the vertical axis
Resistance to the Vf avirulent strain EU-B05
All the 18 HcrVf1 transformants, i.e. six independent transgenic lines of each SPHcrVf1, LPHcrVf1, and PMdRbc HcrVf1, showed heavy sporulation after inoculation with the Vf avirulent strain EU-B05. Sporulation was statistically similar to the level found in non-transgenic ‘Gala’ (susceptible control) (Supplement 1a). The high sporulation levels observed were independent of promoter type or promoter length. In some transgenic lines the sporulation was even more than in ‘Gala’.
On the other hand, 10 out of 13 HcrVf2 transformants showed less or no sporulation and were statistically similar to ‘Santana’ (resistant control) (Supplement 1b). All six independent transgenic lines of PMdRbc HcrVf2 showed no sporulation at all after inoculation with the Vf avirulent strain EU-B05. Among the plants carrying the constructs with the native promoters, three out of four independent transgenic lines of SPHcrVf2 showed less or no sporulation, compared to untransformed ‘Gala’. Only one of the three independent transgenic lines of LPHcrVf2 showed less or no sporulation. Two transformants viz., LPHcrVf2-4 and PMdRbc HcrVf2-12 showed the lowest level of sporulation among all transformants. A view of the symptoms of sporulation as an indication of susceptibility and resistance offered by the respective promoters and genes used in this study is given in Fig. 5.
Sporulation of Vf avirulent monoconidial isolate EU-B05 of V. inaequalis as observed on leaves of HcrVf transgenic lines and of ‘Santana’ (resistant control) and of ‘Gala’ (susceptible control). The difference in the size of the leaves among the different plants with or without sporulation may be attributed to the improper development of leaves due to V. inaequalis growth
Spectrum of resistance
The complete sporulation pattern of different genotypes and transformants after inoculation with six different fungal monoconidial isolates is summarized in the Table 3. All the HcrVf1 transformants showed heavy sporulation for all isolates tested. The sporulation levels of the HcrVf1 transformant did not differ significantly from the sporulation level of the susceptible control ‘Gala’.
In HcrVf2 all transformants, but one showed a sporulation pattern similar to ‘Santana’ (resistant control). A reduced sporulation was observed for the Vf virulent isolate EU-NL05 and heavy sporulation was found for the Vf virulent isolate EU-D42. The exception was LPHcrVf2-16, which a showed similar sporulation level as ‘Gala’ for all the tested fungal isolates. LPHcrVf2-4 was tested only against isolates EU-D42 and NZ 188, and showed a sporulation of isolate EU-D42 (2.8), and complete resistance against isolate NZ 188 (0.0). Transformant PMdRbc HcrVf2-12 was tested against isolates EU-B05, EU-D42 and NZ 188 and showed resistance against EU-B05 (0.0), and NZ 188 (0.0), and sporulation with EU-D42 (2.1). For comparison: the scab resistant cultivar ‘Santana’ showed a sporulation level of 0.37, 0.37 and 3.37 when inoculated with EU-B05, NZ 188 and EU-D42, respectively.
In a graphic presentation of the data of Table 3, including means and standard errors, two distinct groups of plants could be observed for all the isolates except EU-D42 in case of HcrVf2 transformants and cv. ‘Santana’. One group of plants showed sporulation levels similar to (not significantly different from) the sporulation of the susceptible control. The other group of plants showed similar sporulation as the Vf-cultivar ‘Santana’. These two groups of plants were significantly different from each other for all the Vf avirulent isolates used. These two groups can be regarded as susceptible and resistant group respectively. Among the cultivars and transformants there was no significant difference observed for their response to the Vf virulent isolate EU-D42.
Correlation between sporulation and gene expression
The sporulation data obtained from the greenhouse experiment and the gene expression data from the qRT-PCR analysis were used to study their correlation. In HcrVf1 transformants, where no resistance whatsoever was found and all showed high levels of sporulation, no correlation could be established. When all HcrVf2 transformants were taken together, an overall negative correlation was observed i.e. the higher the expression, the lower the sporulation. The correlation co-efficient observed was −0.57 (P = 0.007) between the expression and sporulation. Looking at the HcrVf2 transformants in more detail, it was found that all the PMdRbc HcrVf2 transformants were statistically similar to ‘Santana’ in resistance and were highest in the gene expression. One of the LPHcrVf2 transformants, LPHcrVf2-4, was the highest in expression among the native promoters, and also proved to be statistically similar to ‘Santana’ in resistance. However two SPHcrVf2 transformants (SPHcrVf2-2 and SPHcrVf2-11) were statistically similar to ‘Santana’ in resistance while having lower level of expression of HcrVf2 compared to ‘Santana’ (0.30 and 0.84 times ‘Santana’ respectively). On the other hand SPHcrVf2-15, LPHcrVf2-1, and LPHcrVf2-16 that also had expression levels close to ‘Santana’, showed complete sporulation like untransformed, susceptible cv. ‘Gala’. Nevertheless, the overall correlation was significant.
Discussion
We intended to develop cisgenic cultivars with resistance to apple scab. For the Vf resistance locus, candidate genes had already been isolated and functionally analyzed. The Vf locus comprises of four paralogs namely HcrVf1, HcrVf2, HcrVf3 and HcrVf4 (Vinatzer et al. 2001). HcrVf3 and HcrVf4 were predicted to be truncated and non-functional genes (Xu and Korban 2002). The roles of HcrVf1 and HcrVf2 were investigated first by Malnoy et al. (2008). They transformed Vfa1 (which is the same as HcrVf1) and Vfa2 (which is the same as HcrVf2) with their native promoters of 2 kb in length and their native terminators, to the susceptible cultivars ‘Galaxy’ and ‘McIntosh’. In their hands, Vfa1 and Vfa2 both gave partial resistance to a mixture of isolates (Race 1–Race 5) of V. inaequalis with a reduction in sporulation by 50 and 38% in plants with Vfa1 and Vfa2 respectively. Later, Szankowski et al. (2009) showed complete resistance by inserting HcrVf2 under the regulation of native promoters of 288 and 799 bp length and the nos terminator into susceptible cvs. ‘Gala’ (both 288 and 799 bp promoters) and ‘Elstar’ (only 288 bp promoter). However, using a 115 bp promoter giving an HcrVf2 expression in an ‘Elstar’ transformant similar to the expression in the Vf resistant control cv. ‘Florina’, they found no or only a low level of resistance. They used also a mixture of V. inaequalis isolates for scab inoculation. Here, we checked for both HcrVf1 and HcrVf2 individually their effects on sporulation of apple scab, using promoter lengths at both size classes reported to be functional by Malnoy et al. (2008) and by Szankowski et al. (2009). Moreover in an intragenic approach, a promoter of apple origin, i.e. the small subunit rubisco gene promoter, capable of giving expression at high levels (Schaart et al. 2011) was used. In stead of a mixture of isolates, we used at first a monoconidial isolate and later we determined the effect of the individual HcrVf genes on representatives of the spectrum of Vf virulent and Vf avirulent Venturia isolates.
Regulating HcrVf gene expression
In HcrVf1 transformants, the relative expression of the different events showed considerable variation. In spite of this variation, the long promoter proved to give a significantly higher expression than the short promoter. Expression of LPHcrVf1, regulated by the long promoter of 1,990 bp length, was on average ten times higher than that of SPHcrVf1, representing the short promoter of 312 bp (Supplement 2). This could be an indication for the presence of 5′upstream cis-acting elements having a positive effect on driving expression of HcrVf1. Similar results have been obtained in other crops, e.g. in kidney bean (Phaseolus vulgaris L.), where seed-specific Unknown Seed Protein (USP) promoters, short (637 bp) and long (1,149 bp) were used to direct reporter gene expression (Zakharov et al. 2004). They showed that the long promoter led to three times stronger expression than the short promoter. In a similar study, expression of reporter gene gus in tobacco was studied with a series of deleted promoter fragments of the legumin gene (Bäumelein et al. 1991). They showed that major expression-enhancing cis-elements are present beyond 200 bp upstream from the transcription start site of the legumin gene. In apple, Silfverberg-Dilworth et al. (2005) studied different Vf-promoter fragments by combining them with the gus reporter gene in tobacco and they observed results that seem to be in contrast to the findings reported here in apple, although they also met with considerable variability within their samples. They found that an HcrVf1-derived promoter with a length of 1,200 bp showed lower gus expression than shorter HcrVf1 promoters. They concluded that use of promoter fragments greater than 1 kb should be avoided because of reduced activity in tobacco in their experiment. They obtained similar results by combining the gus reporter gene with the HcrVf2 promoter at different lengths and showed that HcrVf2 with a promoter length of 779 bp showed less expression compared to the gene controlled by a short promoter of 288 bp. However, the same research group found in apple that the HcrVf2 gene with a native promoter of 779 bp was expressed at lower levels as compared to a short promoter of 288 bp (Szankowski et al. 2009). Also here, especially within the short promoter group, variability was high.
In our study, as a consequence of the variability observed among the HcrVf2 events, statistical analysis could not demonstrate a significant difference in expression between LPHcrVf2 and SPHcrVf2, even though expression of LPHcrVf2 was on an average five times higher than that of SPHcrVf2 (Supplement 2).
A striking difference was found in the expression of SPHcrVf2 when we compare our results with those of Szankowski et al. (2009). Our results with a short promoter of 288 bp showed expression levels comparable to ‘Santana’, while they found an approximately 20-fold increase with the 288 bp promoter over their resistant control cultivar ‘Florina’. This could be attributed to the different resistant cultivar that was used as reference but also to the use of a native terminator sequence by us and the nos terminator sequence by Szankowski et al. (2009). The significance of terminator sequences in directing expression was first reported by Dean et al. (1989) and confirmed by Ingelbrecht et al. (1989). Schaart et al. (2011) demonstrated in gus expression studies in tobacco that the apple rubisco terminator gave significantly higher expression than the nos terminator when combined with both the apple rubisco promoter or the CaMV35S promoter.
Scab resistance evaluation and correlation with HcrVf gene expression
Since in the natural situation HcrVf1 and HcrVf2 inherit as one locus, it has not been possible to study the role of the individual HcrVf1 and HcrVf2 genes in conferring resistance to different isolates of apple scab when classically bred Vf cultivars are used. We have developed apple transformants with HcrVf1 under the regulation of their short promoter (SP), long promoter (LP) or the apple rubisco gene promoter (PMdRbc) and HcrVf2 under the influence of SP, LP, or PMdRbc. As far as we know, this is the first study on the resistance spectra of HcrVf1 and HcrVf2 separately. Since cv. ‘Gala’ (susceptible control) does not harbor any known effective resistance gene to V. inaequalis it showed high levels of sporulation by all isolates used (Belfanti et al. 2004; Szankowski et al. 2009). All HcrVf1 transformants also showed similar high sporulation with all isolates, demonstrating complete susceptibility, and behaving similar to untransformed cv. ‘Gala’ (Table 3). This result with HcrVf1 is however in contrast to the result obtained by Malnoy et al. (2008). They inserted Vfa1 (which is the same as HcrVf1) and Vfa2 (which is the same as HcrVf2) under their native promoters of 2 kb length and their 1 kb of native terminator into cvs. ‘Galaxy’ and ‘McIntosh’. They used a mixed inoculum representing races 1–5 and they observed only partial resistance in both cases of HcrVf1 and HcrVf2. The partial resistance was explained by either the high concentration (2.7 × 107 conidia/ml) of fungal inoculum used or to a mutation occurring in the HcrVf1 and HcrVf2 genes during the different steps of the transformation process. As a third possible explanation, the physical separation of HcrVf1 and HcrVf2, which in the natural situation exist together, was suggested as possible explanation for the observed partial resistance.
However, we found that HcrVf1, regardless of the promoter and of the expression level, did not provide any resistance to race 1–5, when used separately as monoconidial isolates and therefore, we conclude that HcrVf1 is not functional against V.inaequalis.
HcrVf2 transformants, also regardless of the promoter used and the expression levels obtained, behaved like the Vf cultivar ‘Santana’ (Table 3) with respect to their response to exposure to the different virulent and avirulent isolates. Similar results were obtained by Szankowski et al. (2009) for the short promoter. Together, this indicated that the resistance from the Vf cluster is caused by HcrVf2, and not by HcrVf1. However, we cannot exclude that the partial resistance observed by Malnoy et al. (2008) could be due to inoculation with a mixture of Venturia isolates.
From the HcrVf2 transformants, LPHcrVf2-16 was completely susceptible to all isolates confirming the results of the previous experiment with EU-B05 only. Presence of the HcrVf2 gene was demonstrated in this line and expression was at a similar level to the expression in cv. ‘Santana’. Possibly, in this line the HcrVf2 insert was mutated in its promoter region, resulting in possible failure of inoculation-induced HcrVf2 upregulation, or, alternatively, a mutation in the coding region may have affected translation or functionality of the HcrVf2 protein.
The two best performing apple transformants which gave very high resistance and showed a relatively high HcrVf2 gene expression are LPHcrVf2-4 and PMdRbc HcrVf2-12. They showed only sporulation with the Vf virulent isolate EU-D42. This indicates that the isolate EU-D42 lacks the avirulence (AVR) protein that is putatively recognized by the HcrVf2 gene. The reason for non-recognition of AVR genes may be either due to mutation or silencing of Vf-AVR gene in the isolate EU-D42. Another explanation could be that the isolate has such AVR genes, but is able to block the downstream defense reaction. Still, the Vf virulent isolate EU-D42 was confirmed to be virulent, also in case of overexpression of HcrVf2. Even the 7- and 163-fold increase in HcrVf2 gene expression as compared to cv. ‘Santana’ could not help in providing resistance against this Vf virulent isolate.
Among all lines and cultivars checked with the spectrum of scab isolates, two clear groups could be distinguished except for isolate EU-D42 (Table 3). It is evident from Table 3 that in general HcrVf1 transformants followed the pattern of untransformed cv ‘Gala’ and HcrVf2 transformants followed the pattern of cv. ‘Santana’. Table 3 also shows the relation between expression of HcrVf genes and sporulation of different isolates. In ‘Gala’ due to the absence of scab resistance genes there was no expression and the sporulation was high irrespective of the isolates. In spite of the high expression of HcrVf1 in transformants SPHcrVf1-12 LPHcrVf1-7 and PMdRbc HcrVf1-9 these plants showed sporulation levels similar to cv. ‘Gala’ irrespective of the isolates tested. So in conclusion, there was no correlation between expression of HcrVf1 and sporulation for any of the isolates tested. For HcrVf2, the picture was also clear, except for transformant LPHcrVf2-16. In cv. ‘Santana’, SPHcrVf2-11, PMdRbc HcrVf2-3, PMdRbc HcrVf2-12, and LPHcrVf2-4 expression of HcrVf2 provided resistance against the Vf avirulent isolates (EU-B05, 1639, US-3 and NZ 188), led to reduced sporulation of Vf virulent isolate EU-NL05 and could not prevent complete sporulation of Vf virulent isolate EU-D42. This is a strong indication that the resistance coming from the Vf cluster is entirely and solely due to HcrVf2 and not due to HcrVf1.
Correlation between T-DNA insert copy number and gene expression
Twenty-six apple transformants were analyzed for transgene copy number through Southern hybridization. Seventeen of them were found to have a single copy inserted and seven carried two T-DNA inserts. No individuals carrying more than two T-DNA copies were obtained. Both types (single copy and two copies) of transformants showed a wide variation in transgene expression and neither a positive nor a negative correlation could be found between copy number and gene expression. In literature, negative correlations between T-DNA insert copy number and expression have been reported due to co-suppression, e.g. in Citrus where a negative correlation between gus gene copy number and GUS activity was found by Cervera et al. (2000). In other cases, no clear correlations could be found (Jones et al. 1985; Zanek et al. 2009; Zeng et al. 2009), as was the case in this study. A correlation did exist between expression and sporulation in HcrVf2 transformants but, as can be seen in supplement 3, expression beyond a certain level did not result in even higher resistance nor in an extension of the resistance spectrum to Vf virulent isolates.
Most variation in the observed expression among different transformation events with the same number of copies is thought to be due to the position effect of the inserted T-DNA (Dean et al. 1988). From two of the LPHcrVf1 transformants, LPHcrVf1-7 and LPHcrVf1-4, LPHcrVf1-7 has a single copy insert and showed a fourfold higher gene expression level than LPHcrVf1-4 which has two transgene copies. In the case of the two PMdRbc HcrVf2 transformants, PMdRbc HcrVf2-11 and PMdRbc HcrVf2-12, PMdRbc HcrVf2-11 has one transgene copy and showed more than twofold lower gene expression than PMdRbc HcrVf2-12 that has two copies.
In conclusion, two genes HcVf1 and HcrVf2 under the regulation of their native promoter (SP and LP cisgenic approach) and the apple rubisco promoter (intragenic approach) were studied for expression and resistance against six isolates of V. inaequalis. It was proven that the resistance provided by the Vf cluster is from HcrVf2. HcrVf1 does not confer resistance against any used isolate. Increasing the HcrVf2 gene expression to high levels did not help in conferring resistance against Vf virulent isolates, but is helpful for resistance against Vf avirulent isolates. From this perspective, the HcrVf2 gene is presently the best choice for development of good resistance against Vf avirulent isolates and can certainly be a good choice for generating both cisgenic and intragenic plants. In order to get even better and more durable resistance we need more resistance genes against apple scab giving a broad spectrum resistance against Vf avirulent and virulent isolates and ways to combine them efficiently and rapidly.
References
Bäumelein H, Boerjan W, Nagy I, Panitz R, Inze D, Wobus U (1991) Upstream sequences regulating legumin gene expression in heterologous transgenic plants. Mol Gen Genet 225:121–128
Belfanti E, Silfverberg-Dilworth E, Tartarini S, Patocchi A, Barbieri M, Zhu J, Vinatzer BA, Gianfranceschi L, Gessler C, Sansavini S (2004) The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc Natl Acad Sci USA 101:886–890
Bus VGM, Rikkerink EHA, Van de Weg WE, Rusholme RL, Gardiner SE, Bassett HCM, Kodde LP, Parisi L, Laurens FND, Meulenbroek EJ, Plummer KM (2005) The Vh2 and Vh4 scab resistance genes in two differential hosts derived from Russian Apple R12740–7A map to the same linkage group of apple. Mol Breeding 15:103–116
Cervera M, Pina JA, Navarro L, Peña L (2000) A broad exploration of transgenic population of citrus: stability of gene expression and phenotype. Theor Appl Genet 100:670–677
Chevalier M, Lespinasse Y, Renaudin S (1991) A microscopy study of different classes of symptoms coded by the Vf gene in apple for resistance to scab (Venturia inaequalis). Plant Pathol 40:249–256
Croxall HE, Gwynne DC, Jenkins JEE (1952) The rapid assessment of apple scab on leaves. Plant Pathol 1:39–41
Dean C, Jones J, Favreau M, Dunsmuir P, Bedbrook J (1988) Influence of flanking sequences on variability in expression levels of an introduced gene in transgenic tobacco plants. Nucleic Acids Res 16:9267–9283
Dean C, Favreau M, Bond-Nutter D, Bedbrook J, Dunsmuir P (1989) Sequences downstream of translation start regulate quantitative expression of two Petunia rbcS genes. Plant Cell 1:201–208
Durel CE, Parisi L, Laurens F, Van de Weg WE, Leibhard R, Jourjon MF (2003) Genetic dissection of partial resistance to race 6 of Venturia inaequalis in apple. Genome 46:224–234
Ellis RJ (1979) The most abundant protein in the world. Trends Biochem Sci 4:241–244
Erdin N, Tartarini S, Broggini GAL, Gennari F, Sansavini S, Gessler C, Patocchi A (2006) Mapping of the apple scab-resistance gene Vb. Genome 49:1238–1245
Espley RV, Hellens RP, Puterill J, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of a MYB transcription factor, MdMYB10. Plant J 49:414–427
Genstat® (2009) Genstat 12 reference manual. VSN International Ltd, Hempstead
Gygax M, Gianfranceschi L, Liebhard R, Kellerhals M, Gessler C, Patocchi A (2004) Molecular markers linked to the apple scab resistance gene Vbj derived from Malus baccata jackii. Theor Appl Genet 109:1702–1709
Hemmat M, Brown SK, Aldwinckle HS, Weeden NF (2003) Identification and mapping of markers for resistance to apple scab from “Antonovka” and “Hansen’s baccata #2”. Acta Hortic 622:153–161
Ingelbrecht LW, Herman LMF, Dekeyser RA, Van Mountagu MC, Depicker AG (1989) Different 3′ end regions strongly influence the level of gene expression in plan cells. Plant Cell 1:671–680
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29:e25
Jacobsen E, Schouten HJ (2008) Cisgenesis, a new tool for traditional plant breeding, should be exempted from the regulation on genetically modified organisms in a step by step approach. Potato Res 51:75–88
Jones JDG, Gilbert DE, Grady KL, Jorgensen RA (1985) T-DNA structure and gene expression in petunia plants transformed by Agrobacterium tumefaciens c58 derivatives. Mol Gen Genet 207:478–485
Lane WD, Bhagwat B, Armstrong JD, Wahlgren S (2003) Apple micrografting protocol to establish transgenic clones on field ready rootstock. HortTechnology 13:641–646
Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation competent Arabidopsis genomic library in Agrobacterium. Nat Biotechnol 9:963–967
Li Z, Hansen JL, Liu Y, Zemetra RS, Berger PH (2004) Using real-time PCR to determine copy number in wheat. Plant Mol Biol Rep 22:179–188
Malnoy M, Xu M, Borejsza-Wysocka E, Korban SS, Aldwinckle HS (2008) Two receptor-like genes, Vfa1 and Vfa2, confer resistance to the fungal pathogen Venturia inaequalis inciting apple scab disease. Mol Plant Microbe Interact 21:448–458
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Outchkourov NS, Peters J, Dong JD, Rademakers W, Jongsma MA (2003) The promoter-terminator of chrysanthemum RbcS1 directs very high expression levels in plants. Planta 216:1003–1012
Parisi L, Lespinasse Y, Guillaumes J, Krüger J (1993) A new race of Venturia inaequalis virulent to apple with resistance due to the Vf gene. Phytopathology 83:533–537
Parisi L, Fouillet V, Schouten HJ, Groenwold R, Laurens F, Didelot F, Evans K, Fischer C, Gennari F, Kemp H, Lateur M, Patocchi A, Thissen J, Tsipouridis C (2004) Variability of the pathogenicity of Venturia inaequalis in Europe. Acta Hortic 663:107–113
Patocchi A, Gianfranceschi L, Gessler C (1999) Towards the map-based cloning of Vf: fine and physical mapping of the Vf region. Theor Appl Genet 99:1012–1017
Patocchi A, Bigler B, Koller B, Kellerhals M, Gessler C (2004) Vr2: a new apple scab resistance gene. Theor Appl Genet 109:1087–1092
Puite KJ, Schaart JG (1996) Genetic modification of the commercial apple cultivars Gala, Golden Delicious and Elstar via an Agrobacterium tumefaciens-mediated transformation method. Plant Sci 119:125–133
Rebrikov DV, Trofimov DY (2006) Real time PCR: a review of approaches to data analysis. Appl Biochem Microbiol 42:455–463
Rommens CM, Haring MA, Swords K, Davies HV, Belknap WR (2007) The intragenic approach as a new extension of traditional plant breeding. Trends Plant Sci 12:397–403
Rouppe Van der Voort J, Janyuka K, Van der Vossen E, Bendahmane A, Mooijman P, klein-Lankhorst R, Stiekema W, Baulcombe D, Bakker J (1999) Tight physical linkage of the nematode resistance gene Gpa2, the virus resistance gene Rx on a single segment introgressed from the wild species Solanum tuberosum subsp. andigena CPC 1673 into cultivated potato. Mol Plant Microbe Interact 12:197–206
Schaart JG, Krens FA, Pelgrom KTB, Mendes O, Rouwendal GJA (2004) Effective production of marker-free transgenic strawberry plant using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotechnol J 2:233–240
Schaart JG, Krens FA, Wolters A-MA, Visser RGF (2010) Chapter 24, Transformation methods for obtaining marker-free genetically modified plants. In: Steward Jr CN, Touraev A, Citovsky V, Tzfira T (Eds) Plant transformation technologies. Wiley-Blackwell, Oxford pp 229–242
Schaart JG, Tinnenbroek IEM, Krens F (2011) Isolation and characterization of strong gene regulatory sequences from apple, Malus × domstica. Tree Genet Genomes 7:135–142
Schouten HJ, Krens FA, Jacobsen E (2006a) Cisgenic plants are similar to traditionally bred plants. EMBO Rep 7:750–753
Schouten HJ, Krens FA, Jacobsen E (2006b) Do cisgenic plants warrant less stringent oversight? Nat Biotechnol 24:753
Silfverberg-Dilworth E, Besse S, Paris R, Belfanti E, Tartarini S, Sansavini S, Patocchi A, Gessler C (2005) Identification of functional apple scab resistance gene promoters. Theor Appl Genet 110:1119–1126
Soriano JM, Joshi SG, Van Kaauwen M, Noordijk Y, Groenwold R, Henken B, Van de Weg WE, Schouten HJ (2009) Identification and mapping of the novel apple scab resistance gene Vd3. Tree Genet Genomes 5:475–482
Southern EM (1975) Detection of specific sequence among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Suzuki Y, Nakabayashi K, Yoshizawa R, Mae T, Makino A (2009) Differences in expression of the RBCS multigene family and Rubisco protein content in various Rice plant tissues at different growth stages. Plant Cell Physiol 50:1851–1855
Szankowski I, Waidmann S, Degenhardt J, Patocchi A, Paris R, Silfverberg-Dilworth E, Broggini G, Gessler C (2009) Highly scab-resistant transgenic apple lines achieved by introgression of HcrVf2 controlled by different native promoter lengths. Tree Genet Genomes 5:349–358
Vinatzer BA, Patocchi A, Gianfranceschi L, Tartarini S, Zhang HB, Gessler C, Sansavini S (2001) Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes co segregating with Vf apple scab resistance. Mol Plant Microbe Interact 14:508–515
Xu M, Korban SS (2002) A cluster of four receptor-like genes resides in the Vf locus that confers resistance to apple scab disease. Genetics 162:1995–2006
Zakharov A, Giesberg M, Hosein F, Meizer M, Müntz K, Saalback I (2004) Seed-specific promoters direct gene expression in non-seed tissue. J Exp Bot 55:1463–1471
Zanek MC, Reys CA, Cervera M, Peña EJ, Velázqez K, Costa N, Plata MI, Grau O, Peña L, García ML (2009) Genetic transformation of sweet orange with the coat protein gene of citrus psorosis virus and evaluation of resistance against the virus. Plant Cell Rep 27:57–66
Zeng F, Zhan Y, Nan N, Xin Y, Qi F, Yang C (2009) Expression of bgt gene in transgenic birch (Betual platyphylla Suk.). Afr J Biotechnol 8:3392–3398
Acknowledgments
We thank Mrs. Iris Tinnenbroek-Capel and Mrs. Linda Kodde (Wageningen UR Plant Breeding, The Netherlands) for their help during the lab experiments, Ms. Bellancile Uzayisenga, M.Sc. student for her help during the greenhouse experiments, and Dr. Chris Maliepard (Wageningen UR Plant Breeding, Wageningen, The Netherlands) for all the help and advice during the statistical analysis of the results. This research was made possible by TransForum, Netherlands. TransForum encourages the necessary sustainable of Dutch agriculture by linking it to its metropolitan environment.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Joshi, S.G., Schaart, J.G., Groenwold, R. et al. Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol Biol 75, 579–591 (2011). https://doi.org/10.1007/s11103-011-9749-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9749-1