Abstract
Apple scab, caused by the fungal pathogen Venturia inaequalis, is one of the most devastating diseases for the apple growing in temperate zones with humid springs and summers. Breeding programs around the world have been able to identify several sources of resistance, the Vf from Malus floribunda 821 being the most frequently used. The appearance of two new races of V. inaequalis (races 6 and 7) in several European countries that are able to overcome the resistance of the Vf gene put in evidence the necessity of the combination of different resistance genes in the same genotype (pyramiding). Here, we report the identification and mapping of a new apple scab resistance gene (Vd3) from the resistant selection “1980-015-25” of the apple breeding program at Plant Research International, The Netherlands. This selection contains also the Vf gene and the novel V25 gene for apple scab resistance. We mapped Vd3 on linkage group 1, 1 cM to the south of Vf in repulsion phase to it. Based on pedigree analysis and resistance tests, it could be deduced that 1980-015-25 had inherited Vd3 from the founder “D3.” This gene provides resistance to the highly virulent EU-NL-24 strain of race 7 of V. inaequalis capable of overcoming the resistance from Vf and Vg.
Similar content being viewed by others
Introduction
Apple scab, caused by the fungal pathogen Venturia inaequalis (Cooke) G.Wint., is one of the most devastating diseases for apples (Malus x domestica Borkh) in temperate zones with humid growing seasons. Most of the commercial apple cultivars are susceptible to the disease, and growers have to spray 20–30 times with fungicides in a season. The use of resistant cultivars could reduce the cost to the growers and may also contribute to a cleaner environment and to a reduction of fungicide residuals on apples for consumers.
The most widely used apple scab resistance gene is Vf from Malus floribunda 821. However, in the meantime, V. inaequalis strains have been detected that are able to overcome the Vf resistance (Parisi et al. 1993). Especially in northwestern Europe, these strains are present and have spread around (Parisi et al. 2006). As a result, several orchards consisting of Vf-cultivars have to be sprayed like orchards with susceptible cultivars (Trapman 2006). For durable resistance, several resistance genes should be accumulated (pyramiding). Fortunately, new loci, which include both major genes and quantitative trait loci (QTLs) that confer resistance to a broad spectrum of V. inaequalis strains, have been discovered in Malus (Calenge et al. 2004; Schmidt and Van de Weg 2005; Gessler et al. 2006; Gardiner et al. 2006). Until now, 11 major apple scab resistance genes have been mapped (Table 1). Some of them are considered as ephemeral resistance genes, acting only against a very narrow spectrum of races of V. inaequalis. Molecular markers linked to these genes are available (Gessler et al. 2006), and in this context, marker-assisted selection could be a useful tool in order to accelerate the breeding programs, for example, selecting the parents for the future crosses.
In this work, we report the identification and mapping of a new qualitative apple scab resistance gene named Vd3 from the selection “1980-015-25” of the apple breeding program of Plant Research International in Wageningen, the Netherlands. This gene provides resistance to the highly virulent EU-NL-24 strain of race 7 of V. inaequalis, which has overcome the resistance from Vf and Vg.
Material and methods
Plant material and DNA extraction
For the mapping of Vd3, we used the population 2000-012C (Table 2). This population is a part of the population 2000-012 comprising 894 individuals and derived from the cross between the scab-resistant selection “1980-015-025” and the susceptible selection “1973-001-041.” In the resistance tests, the cultivars “Elstar,” “Priscilla,” “Gala,” and “Golden Delicious”; the selections “D3,” “1980-015-025,” and “1973-001-041”; and some individuals of the population 2000-012 carrying only Vd3 were used. For pedigree analysis, the apple cultivars Elstar and Priscilla and the selections D3, 1972-010-33, and 1980-015-025 were tested with the simple sequence repeat (SSR) CH-Vf1. DNA extraction was performed from unfolded leaves of apple as described for the Diversity Arrays Technology (DArT) technique (Jaccoud et al. 2001) in http://www.diversityarrays.com.
Evaluation of scab resistance
Scab resistance was evaluated after tunnel tests with mist evaporation, where the four youngest leaves of six replicates of the progeny 2000-012 containing Vd3 only and reference cultivars (Elstar, D3, Priscilla, Gala, Golden Delicious, 1980-015-025, and 1973-001-041) (Table 3) were inoculated with a monoconidial suspension of the nine different races of V. inaequalis (105 conidia/ml) used in the disease tests (Table 3). These isolates belong to the European collection of V. inaequalis from the “Durable Apple Resistance in Europe” project (Lespinasse et al. 2000). Plants were incubated for 48 h at 20°C and 100% relative humidity, and then transferred to a greenhouse with a relative humidity of 85–90%. Disease symptoms were assessed macroscopically after 14–17 days, indicating the presence (susceptible plant) or absence (resistant plant) of sporulation.
The population 2000-012C was inoculated a few weeks after emergence of the young seedlings. For mapping purposes, the strain EU-NL-24 was used. V25 confers resistance to this strain. This hampered mapping of the other resistance gene, i.e., Vd3. Therefore, the 143 plants that contained V25 were discarded based on flanking markers of V25. The remaining 92 plants were used for mapping Vd3. EU-NL-24 is capable of overcoming the Vf gene (Parisi et al 2004) and combines the virulences of races 6 and 7 (Calenge et al. 2004). Disease symptoms were assessed macroscopically after 14–17 days and rated in eight classes indicating the amount of sporulation as follows: class 0, 0% of sporulation; class 1, 1–2% sporulation; class 2, 2–5% sporulation; class 3, 5–10% sporulation; class 4, 10–25% sporulation; class 5, 25–50% sporulation; class 6, 50–75% sporulation; class 7, 75–100% sporulation. This scale is similar to that reported by Durel et al. (2003), which was adapted from Croxall et al. (1952). The two youngest inoculated leaves were scored for sporulation.
DArT markers
DArT markers were produced by Diversity Arrays Technology (Yarralumla, Australia) as described in Wenzl et al. (2004) and Wittenberg et al. (2005).
SSR markers
All of the SSRs in linkage group (LG) 1 available at the High-quality Disease Resistant Apples for a Sustainable Agriculture (HiDRAS) database (http://www.hidras.unimi.it/) were selected. In total, nine SSRs were screened in the population 2000-012C (Table 4). SSR amplifications were performed in a GeneAmp® PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA) in a final volume of 20 μl, containing 75 mM Tris–HCl, pH 8.8; 20 mM (NH4)2SO4; 1.5 mM MgCl2; 0.2 mM of each dNTP; 0.5 μM of fluorescent dye-labeled forward primer (Hex or 6-Fam, Biolegio, Nijmegen, the Netherlands); 0.5 μM of reverse primer; 20 ng of genomic DNA; and 1 U of SuperTaq DNA polymerase (HT Biotechnology, Cambridge, UK) using the following temperature profile: 94°C for 2 min 30 s, then 34 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, finishing with 72°C for 5 min. Samples were analyzed on an ABI PRISM 3700 DNA Analyzer (Applied Biosystems) and scored with GENOTYPER version 3.6 (Applied Biosystems).
Testing of specific markers linked to other scab resistance genes in LG1
RAPD marker P-136
The Va-linked RAPD marker P-136 reported by Hemmat et al. (2003) was also analyzed in the population 2000-012C to test its association with Vd3. RAPD amplification was performed in a GeneAmp® PCR System 9700 thermal cycler (Applied Biosystems) as described in Hemmat et al. (2003). PCR products were separated on 2% agarose gels (Hispanagar, Burgos, Spain).
Vf2ARD marker
The Vf2ARD marker developed by Boudichevskaia et al. (2008) was tested in the population 2000-012C. The PCR was performed in a GeneAmp® PCR System 9700 thermal cycler (Applied Biosystems) as described in Boudichevskaia et al. (2008). PCR products were separated on 1% agarose gels (Hispanagar).
Linkage analysis
The linkage analysis was carried out using JoinMap 4.0 software (Van Ooijen 2006) with the Kosambi mapping function (Kosambi 1944) used to convert recombination units into genetic distances. LGs were established using as threshold a minimum logarithm of odds of 6.0 and a recombination frequency lower than 0.4. The Vd3 gene was mapped as a dominant gene based on the phenotype data (1 for resistance and 0 for susceptibility). The genetic linkage map was constructed for the resistant parent following the “two-way pseudo test-cross” model of analysis (Grattapaglia and Sederoff 1994) and setting a “cross-pollinator” data type.
Results
Evaluation of scab resistance
The population 2000-012C, segregating for Vf, V25, and Vd3, was used to evaluate the resistance against the monocodial strain of V. inaequalis EU-NL-24, capable of overcoming the resistance conferred by the Vf gene. The other strains used in the disease test overcame the resistance conferred by the Vd3 gene (Table 3). The resistance donor was the selection 1980-015-025, which contains the Vf, V25, and Vd3 genes. This selection was heterozygous for the Vd3 trait, and the susceptible parent was homozygous recessive. From 14 to 17 days after inoculation, plants were scored for scab resistance and classified into eight classes based on symptoms (see “Material and methods” section). As the monoconidial strain is virulent to the Vf resistance, two effective major resistance genes were left, i.e., V25 and Vd3. V25 and Vd3 inherited independently. For the purpose of mapping Vd3 precisely, 143 progeny plants carrying the V25 gene were discarded using markers that flanked this gene tightly (unpublished data). The sporulation of the remaining seedlings (N = 92) is depicted in the histogram in Fig. 1. The resistance reaction observed in the plants carrying Vd3 was a hypersensitive pit type and chlorotic reactions.
Based on this histogram, those nonsporulating plants were designated as resistant, while those sporulating plants were designated as susceptible. On the basis of this scoring, 41 out of the 92 F1 individuals were considered resistant and 51 susceptible (Table 2). Segregation of resistance and susceptibility in this cross fit the ratio of 1:1, based on the Chi-square test (χ 2 = 1.08) using a significance level of p value of 0.05, indicating a monogenic inheritance. The hypothesis of two independent major dominant genes involved in the resistance was also tested, but it was rejected due to the high values of the Chi-square test (18.71) (Table 2).
Construction of the LG1 of 1980-015-025
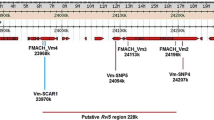
This new gene could be mapped on LG1, closely to Vf, but on the homologous chromosome, so in repulsion to the CH-Vf marker (Vinatzer et al. 2004). For further mapping of Vd3, SSRs in LG1 were tested. Seven out of nine SSRs screened were polymorphic in the mapping population, and five of them were incorporated to the 1980-015-025 map (Table 4). In the case of the DArT markers, out of 234 markers from the resistant parent and absent in the susceptible parent, five were mapped on LG1. Other markers linked to scab resistance genes in LG1 were tested in population 2000-012C. With the first one, the RAPD marker P-136 linked to the Va gene according to Hemmat et al. (2003) should give the band of 700 bp, but this band was not present in the population 2000-012C (data not shown). The second marker was the marker Vf2ARD developed by Boudichevskaia et al. (2008) based on the sequence of HcrVf2 (Vinatzer et al. 2001) in the apple accessions “Antonovka,” “Realka,” and “Discovery.” It was mapped in a similar location to the north of Vf, as reported by these authors (Fig. 2), but the marker was also present in the susceptible parent of our population (data not shown).
Genetic maps of Vd3. a LG1 of 1980-015-025 showing the estimated position of Vf and Va genes according to Gessler et al. (2006). b Map position of Vd3 in LG1. c Map position of Vd3 after removing the phenotype information of the three genotype–phenotype incongruent (GPI) plants
At first, LG1 was created only with SSR and DArT markers (Fig. 2a). This map covers a distance of 58.9 cM comprising 11 loci. The SSRs mapped maintain the colinearity with the Discovery map of Silfverberg-Dilworth et al. (2006) and with the “Florina” map reported by Broggini et al. (2009). Finally, Vd3 was mapped on LG1 as a dominant heterozygous gene, using the resistance data of the segregating population (Fig. 1). In the first location, this gene mapped between the SSR CH-Vf1 and the DArT 67005F17 at a distance of 2.9 cM from the Vf locus (Fig. 2b). After careful observation of the resistant/susceptible phenotype and genotype of the markers around Vd3, it was observed that three progeny plants (one belonging to class 1 and two to class 2) showed genotype–phenotype incongruence (GPI) (Gygax et al. 2004). These plants, classified as susceptible, showed either marker alleles in coupling with the resistance allele. In this case, by removing the resistance data of these plants, Vd3 was mapped again between the markers CH-Vf1 and 67005F17, but the distance to the Vf locus was reduced to 1.0 cM (Fig. 2c). Exclusion of the GPI plants did not change the order of the markers, and the length of the LG1 was only reduced 0.2 cM.
Pedigree analysis of 1980-015-025 and resistance source of Vd3
Figure 3 shows the parents and grandparents of the selection 1980-015-025. The male parent Priscilla was used as donor of the Vf resistance. This could be confirmed with the 159-bp allele from CH-Vf1 linked to HcrVf1 (Vinatzer et al. 2004). This band was only present in the Vf cultivar Priscilla and in 1980-015-025.
As D3 appeared to be completely resistant in the field, this founder was used by the breeders of Plant Research International as a donor of additional resistance. They crossed it with the susceptible cultivar Elstar. Elstar and Priscilla are susceptible to the strain EU-NL-24 (Table 3) and, thus, lack the Vd3 gene. However, D3 is resistant to EU-NL-24 (Table 3). This suggests that Vd3 comes from D3. This could be confirmed with the alleles of the CH-Vf1 SSR, as shown in Fig. 3: Vf cosegregates with the 159-bp band of the SSR, whereas Vd3 cosegretates with the 129-bp band. The latter band originates from the source of resistance D3.
Discussion
Resistance screening
The correct placement of the threshold between resistance and susceptibility is very important to determine the resistance inheritance model and the position of the resistance gene in the linkage map. The selection of the threshold used in this work was due to the degree of sporulation observed from class 1 to class 7 and the absence of V. inaequalis spores in class 0 (Fig. 1). Comparison with the genetic markers indicates that this threshold gives good results, apart from three seedlings with 1–5% of sporulation. This indicates that Vd3 usually provided complete resistance, but 3% of the seedlings showed a low level of sporulation in spite of the presence of Vd3. We did not observe any healthy plants lacking Vd3. This implies that we had no plants that escaped from inoculation.
Linkage map
An accurate phenotyping of the resistance/susceptible data in the progenies is essential for the correct placement of the gene of interest in the linkage map. Two positions are proposed for the Vd3 gene according to the inclusion or exclusion of GPI plants of the analysis (Fig. 2b, c). In both situations, the gene is mapped between the same markers, and the order of the markers in the map is not altered, but in the case of the exclusion of the GPI plants, the distance between Vd3 and the Vf locus was reduced from 2.9 to 1 cM. As was reported by other authors (Patocchi et al. 1999a; Gygax et al. 2004; Erdin et al. 2006), we found difficulties determining the correct position of Vd3, as the phenotypic data were not as precise as the molecular data. During the analysis of the population 2000-012C, we classified three plants (3%) as susceptible, but the flanking marker alleles were in coupling with the resistance. One susceptible plant without Vf showed from 2% to 5% sporulation. As we classified 2–5% sporulation as susceptible and absence of Vd3, this plant would lack both Vf and Vd3. This is the only indication of a recombination between the Vf locus and the Vd3 locus. The flanking markers confirm this recombination event around the Vf locus in this plant. This single plant is responsible for positioning Vd3 just south of Vf, and not at the Vf locus itself or north of Vf. If the sporulation of 2–5% was regarded as resistant and the presence of Vd3 or all of the plants in classes 1 and 2 were discarded, then Vd3 would have been positioned in the same bin as the Vf locus. We conclude that there is evidence that Vd3 is south of Vf, but for a firm proof, additional observations are needed.
Patocchi et al. (1999a), during the fine mapping of the Vf gene, classified about 9% of the plants as resistant, but they showed the alleles in repulsion to Vf in the flanking markers. The exclusion of the phenotype data of these plants was necessary in the correct mapping of the gene as they demonstrated later with the map-based cloning of the Vf gene (Patocchi et al. 1999b). In the mapping of the Vbj gene, Gygax et al. (2004) found 13 resistant plants, out of a population of 173 individuals, that did not have the marker alleles in coupling with the gene and 12 susceptible ones that had them. Finally, Erdin et al. (2006), during the mapping of the Vb gene in the population Golden Delicious × “Hansen’s baccata #2,” identified six GPI plants. After the exclusion of the resistance data of these plants, the tension that they observed in the map disappeared. Patocchi et al. (1999a) suggested two hypotheses to explain this situation; the first one is that these plants are double recombinants, and the second one is that they were wrongly classified. As reported by these authors (Patocchi et al. 1999a; Gygax et al. 2004; Erdin et al. 2006), the presence of double recombinants is quite unlikely because of the distance of the flanking markers in coupling with Vd3. In our case, we expect a frequency of double recombinants of 0.6% and not the 3% observed. The hypothesis of the wrong classification could be more probable supported by the fact that these plants were classified in class 1 (one plant) and class 2 (two plants), which means from 1% to 5% of sporulation. Erdin et al (2006) suggested that the presence of susceptible plants showing the marker alleles in coupling with the resistance could be because of the presence of modifiers that might reduce the effect of the resistance gene.
The presence in a small genomic region of Vd3, the Vf locus composed of 4R (resistance)-genes (HcrVf1, HcrVf2, HcrVf3, and HcrVf4) (Vinatzer et al. 2001), and the Vf2ARD (Boudichevskaia et al. 2008) could indicate the presence of a gene cluster of Vf-like sequences. This is supported by the fact that Broggini et al. (2009) found two SSRs developed from BAC clones containing Vf-like sequences in the vicinity of the Vf locus. Another cluster of R-genes against V. inaequalis was found by Bus et al. (2004) in LG2. This cluster comprises four major genes, as well as several race-specific QTLs (Bus et al. 2004). This grouping of R genes in clusters is found frequently in plants. The biological reason could be the generation of novel resistances through the unequal crossover between different genes (Hammond-Kosack and Jones 1997).
Vd3 is a new apple scab resistance gene
The genetic position of Vd3 in LG1 permitted us to show that Vd3 is novel compared to all of the previously identified resistance genes against V. inaequalis, including Vf, Va, and Vf2ARD genes (Patocchi et al. 1999a; Hemmat et al. 2003; Boudichevskaia et al. 2008). In the case of the Vf gene, the resistance spectra demonstrated that Vd3 is not Vf. While the Vf gene has a wide resistance spectrum conferring resistance to the V. inaequalis races 1 to 5 (Mc Hardy 1996), in our disease tests, Vd3 only confers resistance to the EU-NL-24 strain (race 7) capable of overcoming the resistance provided by Vf (Table 3).
Regarding the Va gene, although also mapped in LG1, the position is different to that reported for Vd3. Gessler et al. (2006) reported the positions of the Va gene mapped by Zini (2005), who used the cultivar “Freedom” as the resistance donor. This cultivar is carrying the Vf gene and another one coming from an unspecified Antonovka clone. In the two locations reported, the Va gene is outside the interval between the SSRs Hi12c02 and CH05g08 and, in both cases, at a distance of about 25 cM from Vf (Zini 2005). This distance is in agreement with that reported previously by Hemmat et al. (2003) using as resistance donor “Antonovka PI 172633.” On the contrary, Vd3 is close to Vf (Fig. 2) and is within the region flanked by those SSRs. Using the RAPD marker P-136 described by Hemmat et al. (2003), no results were obtained (data not shown). The band of 700 bp reported by these authors as linked to the Va gene was not present in the population 2000-012C. Therefore, it was not possible to test the association between P-136 and Vd3. Moreover, the reaction types of both genes indicate that they are not the same, as Va induces a hypersensitive pit-type reaction (Dayton and Williams 1968), whereas Vd3 induces a chlorosis reaction too. Finally, taking into account the pedigree analysis showed in Fig. 3, we can say that the source of Vd3 (D3) does not come from an Antonovka accession. The 129-bp allele coming from D3 does not occur in the Antonovka accessions tested by Vinatzer et al. (2004) or in Antonovka 34.16 (unpublished data from HiDRAS database).
Recently, Boudichevskaia et al. (2008) reported the mapping of sequences homologous to the Vf genes identified by Vinatzer et al. (2001) (HcrVf genes). The candidate gene Vf2ARD, developed on the basis of the divergences in the sequence of the C1 subdomain of the HcrVf2 gene present in the apple accessions Antonovka, Realka, and Discovery, was mapped to the north of the Vf locus at a distance of 1.9 cM from the marker CH-Vf1 (Boudichevskaia et al. 2008). Using the primers developed by these authors, we mapped the Vf2ARD gene in a similar location but at a distance between 2.5 and 3 cM, depending on the map (Fig. 2). So Vd3, which is south of the Vf locus, is located at a distance of about 5 cM from Vf2ARD. Moreover, this marker was also present and mapped in the susceptible parent of our population (data not shown). So, we can discard that Vd3 and Vf2ARD were the same gene.
Bénaouf and Parisi (2000) detected in the cross Golden Delicious × M. floribunda 821, in addition to the Vf resistance, another locus for resistance descending from M. floribunda 821. They named it Vfh, but they did not map it. According to Parisi et al. (2004), EU-NL-24 can sporulate on M. floribunda 821. This indicates that the Vfh gene is not the same as the Vd3 gene.
Calenge et al. (2004) mapped QTLs for scab resistance using different strains of V. inaequalis. They also used EU-NL-24. The QTLs for EU-NL-24 were not mapped in the vicinity of the Vf locus in the progeny of the cross derived from Discovery, an English cultivar partially resistant to V. inaequalis, and “TN10-8,” a partially resistant hybrid derived from “Schmidt’s Antonovka PI 172632.”
In conclusion, we present in this work a new apple scab resistance gene that is only 1 cM south of the Vf locus. The V. inaequalis strain EU-NL-24 is virulent to Vf but avirulent to Vd3 cultivars. However, Vd3 has not been effective against the majority of other V. inaequalis strains we used in our disease tests. The Vd3 gene is south of the Vf locus, in contrast to previously mapped HcrVf-like sequences.
References
Bénaouf G, Parisi L (2000) Genetics of host–pathogen relationships between Venturia inaequalis races 6 and 7 and Malus species. Phytopathology 90:236–242
Boudichevskaia A, Flachowsky H, Dunemann F (2008) Identification and molecular analysis of candidate genes homologous to HcrVf genes for scab resistance in apple. Plant Breed. doi:10.1111/j.1439-0523.2008.01537.x
Broggini GAL, Galli P, Parravicini G, Gianfranceschi L, Gessler C, Patocchi A (2009) HcrVf2 paralogs are present on linkage groups 1 and 6 Malus. Genome 52:129–138
Bus V, van de Weg WE, Durel CE, Gessler C, Calenge F, Parisi L, Rikkerink E, Gardiner S, Patocchi A, Meulenbroek M, Schouten H, Laurens F (2004) Delineation of a scab resistance gene cluster on linkage group 2 of apple. Acta Hort 663:57–62
Bus VGM, Rikkerink EHA, van de Weg WE, Rusholme RL Gardiner SE, Bassett HCM et al (2005a) The Vh2 and Vh4 scab resistance genes in two differential hosts derived from Russian Apple R12740-7A map to the same linkage Group of Apple. Mol Breed 15:103–116
Bus VGM, Laurens FND, van de Weg WE, Rusholme RL, Rikkerink EHA, Gardiner SE et al (2005b) The Vh8 locus of a new gene-for-gene interaction between Venturia inaequalis and the wild Apple Malus sieversii is closely linked to the Vh2 locus in Malus pumila. New Phytol 166:1035–1049
Calenge F, Faure A, Goerre M, Gebhardt C, Van de Weg WE, Parisi L, Durel CE (2004) Quantitative trait loci (QTL) analysis reveals both broad-spectrum and isolate-specific QTL for scab resistance in an apple progeny challenged with eight isolates of Venturia inaequalis. Phytopathology 94:370–379
Croxall HE, Gwynne DC, Jenkins JEE (1952) The rapid assessment of apple scab on leaves. Plant Pathol 1:39–41
Dayton DF, Williams EB (1968) Independent genes in Malus for resistance to Venturia inaequalis. Proc Am Soc Hort Sci 92:89–93
Durel CE, van de Weg WE, Venisse JS, Parisi L (2000) Localization of a major gene for apple scab resistance on the European genetic map of the Prima x Fiesta cross. OILB/WPRS Bull 23:245–248
Durel CE, Parisi L, Laurens F, van de Wed WE, Liebhard R, Jourjon MF (2003) Genetic dissection of partial resistance to race 6 of Venturia inaequalis in apple. Genome 46:224–234
Erdin N, Tartarini S, Broggini GAL, Gennari F, Sansavini S, Gessler C, Patocchi A (2006) Mapping of the apple scab resistance gene Vb. Genome 49:1238–1245
Gardiner SE, Bus VGM, Rusholme RL, Chagné D, Rikkerink EHA (2006) Apple. In: Kole C (eds) Genome mapping and molecular breeding. Fruit and nuts. vol. 4. Springer, Berlin, pp 2–62
Gessler C, Patocchi A, Sansavini S, Tartarini S, Gianfranceschi L (2006) Venturia inaequalis resistance in apple. Crit Rev Plant Sci 25:473–503
Grattapaglia D, Sederoff RR (1994) Genetic linkage maps of Eucalyptus grandis and E. urophylla using a pseudotest-cross strategy and RAPD markers. Genetics 137:1121–1137
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus x domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94:249–254
Gygax M, Gianfranceschi L, Liebhard R, Kellerhals M, Gessler C, Patocchi A (2004) Molecular markers linked to the apple scab resistance gene Vbj derived from Malus baccata jackii. Theor Appl Genet 109:1702–1717
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Hemmat M, Brown SK, Aldwinckle HS, Weeden NF (2003) Identification and mapping of markers for resistance to Apple scab from “Antonovka” and “Hansen”s baccata #2”. Acta Hort 622:153–161
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity Arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29:e25
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugenics 12:172–175
Lespinasse Y, Durel CE, Parisi L, Laurens F, Chevalier M, Pinet C (2000) A European project: D.A.R.E.—Durable apple resistance in Europe. Acta Hort 538:197–200
Liebhard R, Gianfrancesschi L, Koller B, Ryder CD, Tarchini R, van de Weg E, Gessler C (2002) Development and characterization of 140 new microsatellites in apple (Malus x domestica Borkh.). Mol Breed 10:217–241
Maliepaard C, Alston FH, van Arkel G, Brown LM, Chevreau E, Dunemann F et al (1998) Aligning male and female linkage maps of Apple (Malus pumila Mill.) using multi-allelic markers. Theor Appl Genet 97:60–73
Mc Hardy WE (1996) Inheritance of resistance to Venturia inaequalis. In: Apple scab, biology, epidemiology and management. APS, St. Paul, pp 61–103
Parisi L, Lespinasse Y, Guillaumes J, Kruger J (1993) A new race of Venturia inaequalis virulent to apples with resistance due to the Vf gene. Phytopathology 83:533–537
Parisi L, Fouillet V, Schouten HJ, Groenwold R, Laurens F, Didelot F, Evans K, Fischer C, Gennari F, Kemp H, Lateur M, Patocchi A, Thissen J, Tsipouridis C (2004) Variability of the pathogenicity of Venturia inaequalis in Europe. Acta Hort 663:107–114
Parisi L, Laurens F, Didelot F, Evans K, Fischer C, Fouillet V, Gennari F, Kemp H, Lateur M, Patocchi A, Schouten HJ, Tsipouridis C (2006) Geographical distribution of Venturia inaequalis strains virulent to the Vf gene in Europe. Bulletin-OILB/SROP 29:49–52
Patocchi A, Gianfranceschi L, Gessler C (1999a) Towards the map-based cloning of Vf: fine and physical mapping of the Vf region. Theor Appl Genet 99:1012–1017
Patocchi A, Vinatzer BA, Gianfranceschi L, Tartarini S, Zhang HB, Sansavini S, Gessler C (1999b) Construction of a 550Kb BAC contig spanning the genomic region containing the apple scab resistance gene Vf. Mol Gen Genet 262:884–891
Patocchi A, Bigler B, Koller B, Kellerhals M, Gessler C (2004) Vr2: A new apple scab resistance gene. Theor Appl Genet 109:1087–1092
Patocchi A, Walser M, Tartarini S, Broggini GAL, Gennari F, Sansavini S, Gessler C (2005) Identification by genome scanning approach (GSA) of a microsatellite tightly associated with the apple scab resistance gene Vm. Genome 48:630–636
Silfverberg-Dilworth E, Matasci CL, van de Weg WE, Van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel CE, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A (2006) Microsatellite markers spanning the apple (Malus x domestica Borkh.) genome. Tree Genet Genom 2:202–224
Schmidt H, van de Weg WE (2005) Breeding. In: Tromp J, Webster AD, Wertheim SJ (eds) Fundamentals of temperate zone tree fruit production. Backhuys, Leiden, pp 136–155
Tartarini S, Gennari F, Pratesi D, Palazzetti C, Sansavini S, Parisi L et al (2004) Characterization and genetic mapping of a major scab resistance gene from the old Italian apple cultivar ‘Durello di Forí’. Acta Hort 663:129–133
Trapman M (2006) Resistance management in Vf resistant organic apple orchards. Bulletin OILB/SROP 29:253–257
Van Ooijen JW (2006) JoinMap®4.0, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Vinatzer BA, Patocchi A, Gianfranceschi L, Tartarini S, Zhang HB, Gessler C, Sansvini S (2001) Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. MPMI 14:508–515
Vinatzer BA, Patocchi A, Tartarini S, Gianfranceschi L, Sansvini S, Gessler C (2004) Isolation of two microsatellite markers from BAC clones of the Vf scab resistance region and molecular characterization of scab-resistant accessions in Malus germplasm. Plant Breed 123:321–326
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A (2004) Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci U S A 101:9915–9920
Wittenberg AHJ, van der Lee T, Cayla C, Kilian A, Visser RGF, Schouten HJ (2005) Validation of the high-throughput marker technology DArT using the model plant Arabidopsis thaliana. Mol Gen Genom 274:30–39
Zini E (2005) Costruzione di una mappa di associazione della popolazione di melo ‘Golden Delicious’ x ‘Freedom’ e caratterizzazione del gene di resistenza Va a ticchiolatura. Ph.D. thesis, DCA-BO, Italy, 126 p
Acknowledgements
This research was supported by Transforum, The Netherlands. JMS was funded by a Postdoctoral contract from the Fundación Española para la Ciencia y la Tecnología (FECYT), Spain. SGJ was funded by Transforum, The Netherlands.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: A. Dandekar
JMS and SGJ contributed equally to this work
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Soriano, J., Joshi, S., van Kaauwen, M. et al. Identification and mapping of the novel apple scab resistance gene Vd3 . Tree Genetics & Genomes 5, 475–482 (2009). https://doi.org/10.1007/s11295-009-0201-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-009-0201-5