Abstract
Chloroplast genome diversity in cork oak (Quercus suber) is characterised by the occurrence of haplotypes that are akin to those found in other Mediterranean oak species, particularly in Q. ilex and Q. rotundifolia, suggesting the possible presence of an introgressed chloroplast lineage. To further investigate this pattern, we reconstructed chloroplast haplotypes by sequencing four chloroplast markers (cpDNA), sampled across 181 individuals and 10 taxa. Our analyses resulted in the identification of two diversified chloroplast haplogroups in Q. suber, corresponding to a geographically widespread lineage and an Afro-Iberian lineage. Time-calibrated phylogenetic analyses of cpDNA point to a Miocene origin of the two haplogroups in Q. suber, suggesting that the Afro-Iberian lineage was present in cork oak before the onset of glaciation periods. The persistence of the two haplogroups in the western part of the species distribution range may be a consequence of either ancient introgression events or chloroplast lineage sorting, combined with different fixation in refugia through glaciation periods. Our results provide a comprehensive insight on the origins of chloroplast diversity in these ecologically and economically important Mediterranean oaks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cork oak (Quercus suber L., Fagaceae) is an evergreen tree species native to the western Mediterranean region where it has considerable economic importance (Aronson et al. 2009; Vessella et al. 2017). Its geographical distribution has been shaped by glaciation cycles (López de Heredia et al. 2007a; Vessella et al. 2015), similarly to other European tree species (Bagnoli et al. 2016; de Dato et al. 2020; Petit et al. 2002). Quercus suber is not only a dominant climax forest species in many natural settings across its distribution range, but also a vital element in managed tree-grassland agro-systems known as “montado” or “dehesa,” together with Q. rotundifolia Lam. These agro-systems are the main source of commercial cork but also provide grazing grounds for livestock (Rolo and Moreno 2019) and harbour significant levels of biodiversity (Lopez-Sanchez et al. 2016).
Quercus suber has a significant regional importance for the primary sector, and has been the subject of numerous studies, for example on population structure (Pina-Martins et al. 2019; Ramírez-Valiente et al. 2010; Soto et al. 2007; Vanhove et al. 2021), adaptation (Modesto et al. 2014; Pina-Martins et al. 2019; Vanhove et al. 2021), hybridisation (Belahbib et al. 2001; Burgarella et al. 2009; López de Heredia et al. 2020; Staudt et al. 2004) and biogeography (López‐de‐Heredia et al. 2005, 2007a; Lumaret et al. 2005; Magri et al. 2007; Simeone et al. 2018; Vila-Viçosa et al. 2020).
Intraspecific genetic diversity in Q. suber can be considered moderate to low and reflects a weak structure along a longitudinal axis (Pina-Martins et al. 2019), but specific regions, in particular along the southern margins of its distribution range, are known to harbour higher levels of genetic diversity (Sousa et al. 2022). Genetic studies in Q. suber have analysed both nuclear data (Burgarella et al. 2009; López de Heredia et al. 2020; Pina-Martins et al. 2019; Vanhove et al. 2021) and chloroplast data (cpDNA), which can be informative in population-level studies of terrestrial plant groups. The chloroplast genome is non-recombinant, usually maternally inherited and haploid, meaning that chloroplast DNA variants, or haplotypes, should become more rapidly fixed within populations, i.e. have a shorter coalescence time, compared to nuclear (diploid) allelic variants (Mariac et al. 2014; Petit and Vendramin 2007). Thus, chloroplast genomic data may provide insights on recent demographic and hybridisation events (López de Heredia et al. 2020; Pham et al. 2017).
Chloroplast haplotypes in Q. suber have been investigated using RFLP and PCR–RFLP markers (Jiménez et al. 2004; López‐de‐Heredia et al. 2005, 2007a, b; Lumaret et al. 2005; Lumaret and Jabbour-Zahab 2009), microsatellites (Magri et al. 2007) and sequence data (Belahbib et al. 2001; López de Heredia et al. 2020; Simeone et al. 2018). The different types of data have shown the presence of two or more haplogroups, which have been interpreted as the result of introgression events, possibly with adaptive influence, between Q. suber and other oak species (Jiménez et al. 2004; Lumaret and Jabbour-Zahab 2009; López de Heredia et al. 2017), as a consequence of isolation and subsequent expansion from glacial refugia (López de Heredia et al. 2007a; Lumaret et al. 2005) or as a product of Miocene plate tectonics (Magri et al. 2007). Reconstructing haplotype patterns from multiple chloroplast sequences and sampling broadly across the distribution range of Q. suber may bring new insights and enable a stronger resolution of haplotype diversity and distribution in this species, compared to earlier inferences made from RFLPs, microsatellites and single chloroplast regions. The main questions to address are whether chloroplast haplotype diversity in Q. suber is geographically structured, and whether it reflects ancient diversification or was instead shaped mostly by hybridisation events with other Quercus L. species. In addition, haplotype sequence data analysis may show which regions harbour the most genetic diversity in the chloroplast genome.
Here we expand the data set of Costa et al. (2011) based on three chloroplast intergenic regions (trnL-F, trnS-psbC and trnH-psbA) by sampling additional populations and by including the matK gene, which is considered a reliable marker for retrieving chloroplast phylogenies (Hochbach et al. 2018). The matK gene has been proposed as a barcode marker for plants (CBOL Plant Working Group. 2009) and has indeed been tested as a barcode marker for genus Quercus (Piredda et al. 2011; Simeone et al. 2013). We conduct diversity and phylogenetic analyses on our expanded dataset with the objective of generating a detailed characterisation of chloroplast diversity and its geographical distribution across the distribution range of Q.suber. We compare our results with earlier analyses of Q. suber cpDNA and use a time-calibrated analysis to elaborate on the origin of Q. suber chloroplast genome diversity.
Materials and methods
Sampling and DNA sequencing
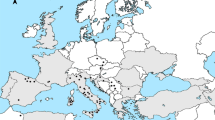
Samples of Quercus suber (subgenus Cerris, sect. Cerris) originated from 26 sites representing the entire natural range of the species. A map showing all sampling sites and the natural distribution of Q. suber was generated based on Caudullo et al. (2017) (Fig. 1).
Diversity measures were estimated at a regional level by combining sites of each country. Leaf material was sampled in situ from natural stands in eight sites in Portugal (Gerês, Serra da Estrela, Serra de São Mamede, Serra da Arrábida, Serra de Monchique, Serra do Buçaco, Azeitão and Serra de Sintra). The remaining samples (Portugal: São Brás de Alportel; Spain: Cataluña, Montes de Toledo, Haza del Lino, Sierra de Aracena, Sierra Morena, Sierra de Guadarrama; Italy: Puglia, Lazio, Sicily; France: Var, Landes, Corsica; Algeria: Forêt des Guerbès; Tunisia: Mekna, Fernana; Morocco: Taza, Kenitra) were obtained from a cork oak provenance trial (FAIR I CT 95 0202) established in 1998 at Herdade Monte da Fava (Santiago do Cacém, Portugal; 8°7′ W, 38°00′ N) as part of the European Forest Genetic Resources Programme (EUFORGEN; Varela 2003). Samples from nine additional Quercus species (subgenus Cerris, sect. Ilex: Q. coccifera L., Q. ilex L., Q. rotundifolia; sect. Cerris: Q. cerris L.; subgenus Quercus, sect. Quercus: Q. canariensis Willd., Q. faginea Lam., Q. lusitanica Lam., Q. pyrenaica Willd.; sect. Lobatae: Q. rubra L.) and from an outgroup species in the Fagaceae (Castanea crenata Siebold & Zucc.) were also sampled. Quercus rotundifolia is often considered a subspecies of Q. ilex (e.g. López de Heredia et al. 2007a). Here we follow the classification of the Plants of the World index (https://powo.science.kew.org) that places Quercus ilex subsp. rotundifolia (Lam.) O.Schwarz ex Tab.Morais in synonymy under Q. rotundifolia Lam. All these oak species have native distribution ranges that overlap with the natural distribution of Q. suber, except for Q. rubra, which is introduced in Europe. All leaf material, corresponding to 181 samples, was stored at −80 °C until DNA extraction. Sampled taxa and sampling sites, with the corresponding site codes, are presented in Table 1.
Leaf tissue was manually ground using liquid nitrogen and genomic DNA was extracted with the DNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. Four chloroplast markers (intergenic spacer regions trnL-F, trnS-psbC, trnH-psbA and the matK gene, in part) were amplified by PCR in a final volume of 25 μL with the following conditions: denaturation step at 94 °C (5’’); 30 cycles of: denaturation at 94 °C (20’’), annealing at 65 °C (for cpDNA intergenic spacers) or 55 °C (for matK) (30’’), extension at 72 °C (40’’); final extension step at 72 °C for 7 min. Primer pairs were obtained from earlier publications (matK: Cuénoud et al. 2002; trnH-psbA: Kress et al. 2005; trnL-F: Taberlet et al. 1998; trnS-psbC: Nishizawa & Watano 2000). Amplified PCR products were verified by gel electrophoresis and purified with SureClean (Bioline). Sequencing of PCR products was done using the BigDye v3.1 chemistry (Applied Biossystems,USA), in house, on an ABI prism 310 automated sequencer, and by outsourcing to Macrogen (South Korea). Sequences were edited with Sequencher v4.0.5 (Gene Codes Corporation).
Alignment, diversity estimates and phylogenetic analyses
Four individual matrices (matK, trnH-psbA, trnL-F, trnS-psbC) were aligned using the software MAFFT v. 7.2 (Katoh and Standley 2013), using a gap penalty of 1 and a maximum of 10 iterative refinements (mafft –op 1.0 –maxiterate 10). The four chloroplast marker alignments were concatenated using the script catfasta2phyml.pl (https://github.com/nylander/catfasta2phyml).
The package “pegas” (v. 1.1; Paradis 2010) implemented in R v. 3.6.3 (R core Team 2013) was used to estimate the number of haplotypes, haplotype and nucleotide diversity, and to test for demographic change using Tajima’s D (Tajima 1989) and R2 (Ramos-Onsins and Rozas 2002).
Maximum-parsimony (MP) trees were inferred from the cpDNA alignment using the program TNT v 1.1 (Goloboff et al. 2008) and a “traditional search” on the Phylogeny.fr server (http://www.phylogeny.fr/). Branch support was estimated from a standard bootstrapping with 1000 replicates. Substitution models were inferred for the concatenated cpDNA alignment using jModelTest v. 2.1 (Darriba et al. 2012) with three substitution schemes and Akaike Information Criterion (AIC) calculations. A maximum-likelihood (ML) full bootstrap analysis of each alignment, with 1000 replicates, was run on RaxML v. 8.2.4 (Stamatakis 2014) using the best-fitting models inferred with AIC (concatenated matrix: GTR) and the gamma model of rate heterogeneity.
A reduced cpDNA alignment with only one sequence per haplotype (46 sequences) was analysed on BEAST v 2.6.7 (Bouckaert et al.2014), under the GTR substitution model and a discrete four category gamma model of site rate heterogeneity. A prior was defined for the monophyly of genus Quercus. A lognormal prior on the TMRCA for the genus was set with a mean in real space of 56E6 (Hipp et al. 2020) and a stdev of 0.05. A relaxed lognormal clock model was applied, with a lognormal prior for ucldmean (mean [real space] = 1.8E-9; stdev = 0.05; adapted from Sousa et al. 2014) and a gamma prior for ucldstdev (α = 0.5396, β = 0.3819). The Yule model was chosen as the tree prior, and a gamma prior (alpha = 0.001, beta = 1000) was used for “birthrate”. The analysis ran for 100E6 MCMC generations on the CIPRES Science gateway (Miller et al. 2011). The run was validated using Tracer v 1.7.2., and all parameters had ESS > 200 after a 10% burnin. A maximum clade credibility tree and branch support values were obtained with TreeAnnotator v 2.6.7.
A haplotype parsimony network was built from the concatenated cpDNA data set with the R package “pegas” and the “haploNet” function, which uses the uncorrected P or Hamming distance and pairwise deletion of missing data.
Results
Analyses of cpDNA sequence data
The concatenated matrix of cpDNA from 181 samples has 2006 base-pairs (bp) in length (matK: 897 bp; trnH-psbA: 478 bp; trnL-F: 381 bp; trnS-psbC: 250 bp), of which 46 are parsimony-informative sites (matK:12; trnH-psbA:18; trnL-F: 8; trnS-psbC:8), and contains 17 indels corresponding to 52 positions with gaps. The largest indel has 11 bp, and 7 indels have 1 bp.
Indels were not coded prior to analyses, i.e. gaps were considered as missing data. This data set, that includes ten sampled Quercus lineages and the outgroup, comprised 46 haplotypes. Among the 146 Q. suber samples, 29 haplotypes were found. Of these, nine (31%) were singletons, i.e. occurred in a single sample. The most frequent haplotype (XVIII) was found in 25 samples from Spain, France and Italy. The second most frequent haplotype (XXII) was found in 23 samples from Portugal. Haplotype II was found to be shared by Q. canariensis, Q. faginea and Q. pyrenaica, which is in accordance with earlier findings (Petit et al. 2002). The full list of samples and corresponding haplotypes is presented in Online Resource 1.
Results of haplotype diversity estimates for Q. suber are presented in Table 2.
The countries with most haplotypes found were Portugal (12), Spain (6) and Italy (5). Haplotype diversity ranged from 0 (Tunisia) to 0.83 (Portugal). The highest nucleotide diversity was found in Morocco (0.005). Tajimas’D statistic was negative in all sites except for Portugal, with significant values in Algeria, France and Italy, whereas the lowest significant values of the R2 test were found in France and Italy.
In parsimony analyses of the complete data set of 181 individuals, 20 trees were retained after 157,888,177 rearrangements. The complete maximum parsimony consensus tree is shown in Online Resource 2, and a reduced MP consensus tree inferred from one sequence of each haplotype is shown in Fig. 2.
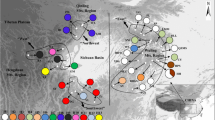
Phylogenetic reconstruction of Quercus suber haplotypes under parsimony. Tree showing the phylogenetic reconstruction produced by a parsimony analysis of the chloroplast sequence data using TNT. This simplified tree is derived from the strict consensus cladogram of 20 most parsimonious trees (Online Resource 2) by merging all tips corresponding to the same haplotype (46 haplotypes). Haplotype numbers are presented in roman numerals. Tip labels show full acronyms for Q. suber, Q. rotundifolia and Q. ilex samples but are shortened for other taxa. Q. suber haplotypes are highlighted in green (suber I) and orange (suber II). Branches with bootstrap support equal or greater than 80 in at least one of the analyses (maximum parsimony and maximum likelihood) are marked with*
Taxa belonging to section Quercus, namely Q. canariensis (QCA), Q. faginea (QFA), Q. pyrenaica (QPY), which share the same haplotype (II), and Q. lusitanica (QLU), form a fully supported clade (BS = 100). The placement of Q. rubra (QRU) as sister to the remainder of the ingroup is also supported (BS = 100), as is the grouping of the two Q. coccifera (QCO) haplotypes. Samples belonging to Q. suber are recovered in two distinct clades: a supported clade (BS = 88 in MP analyses) comprising solely Q. suber lineages from all countries and corresponding to 13 haplotypes (henceforth referred to as the suber I group); an unsupported clade comprising Q. suber samples from Portugal, Spain, Morocco and Algeria, corresponding to 16 haplotypes (henceforth referred to as the suber II group), in which Q. ilex haplotypes appear nested in Q. suber lineages. Relationships among Q. suber haplotypes are largely unresolved, particularly within the suber I group. The two most frequent haplotypes (XVIII, XXII) are included in the suber I clade. Relationships between haplotypes from different taxa are not supported, except for the placement of Q. cerris (QCE) as sister to the suber I group (BS = 92 in MP analyses).
The time-calibrated analysis of the reduced cpDNA alignment using BEAST (Fig. 3) also recovers two Q. suber clades, both with full support (PP = 1.0), and Q. cerris fully supported as sister to the suber I group. A clade composed of three Q. rotundifolia haplotypes (XII, XIII, XIV) and the two Q. coccifera haplotypes (IV, V) is also fully supported. Full support was also obtained for the group comprising all Quercus haplotypes except Q. rubra and Q. rotundifolia haplotype XI.
Time-calibrated phylogenetic reconstruction of Quercus suber haplotypes. Tree showing the phylogenetic reconstruction produced by a time-calibrated analysis of the 46 haplotypes in BEAST2. The inferred tree is scaled to geological time in units of million years (Myr). Branch support values represent posterior probabilities. Haplotype numbers are presented in roman numerals. Q. suber haplotypes are highlighted in green (suber I) and orange (suber II). Pli Pliocene; Ple Pleistocene
Branches between the two suber haplogroups are mostly unsupported. Node height estimates place the root of genus Quercus at 55.25 Myr (height 95% HPD: 49,804,501.61; 60,605,566.97). The split between Q. cerris and Q. suber is estimated at 21.1 Myr (height 95% HPD: 8,957,266.84; 35,020,741.65). The age of the suber I group is estimated at 16.49 Myr (height 95% HPD: 6,483,671.34; 28,170,912.6), whereas the age of the suber II group is estimated to be 23.24 Myr (height 95% HPD: 11,615,111.29; 36,610,615.62).
The parsimony haplotype network built with the uncorrected P distance and showing the different taxa is presented in Fig. 4.
Haplotype parsimony network. Statistical parsimony network (TCS) of haplotypes constructed from the chloroplast sequence data set using the R package “pegas”. Haplotype numbers are presented in roman numerals. Quercus suber haplotypes are highlighted in green (suber I) and orange (suber II). Dashes on each link represent evolutionary steps between haplotypes
Quercus suber haplotypes appear divided into two groups, separated by intermediate Q. rotundifolia haplotypes and the Q. cerris haplotype. The suber I group, comprising 13 Q. suber haplotypes, is separated from the Q. cerris haplotype by seven evolutionary steps (mutations). The suber II group, comprising 12 Iberian and north African Q. suber haplotypes and the Q. ilex haplotype, is separated from Q. rotundifolia by seven steps. Haplotypes in both suber groups are separated by 1–3 steps, except in one case in the suber II group, where six steps separate two of the haplotypes. Six steps separate a group formed by three Q. rotundifolia haplotypes and the two Q. coccifera haplotypes, the latter at a distance of seven steps from the closest Q. rotundifolia haplotype.
Discussion
Our analyses agree with earlier findings that revealed the existence of two main chloroplast haplogroups in Q. suber (Jiménez et al. 2004; López de Heredia et al. 2005, 2007a, b, 2020; Lumaret et al. 2005; Lumaret and Jabbour-Zahab 2009; Simeone et al. 2018). We identify a widespread group, suber I, and a western group, suber II, which is only present in the Iberian Peninsula and NW Africa. Group suber I appears as sister to Q. cerris in all trees, which is in accordance with the taxonomic treatment and with the phylogeny of genus Quercus, that place Q. suber in section Cerris (Denk et al. 2017; Hipp et al. 2020; Hubert et al. 2014; Zhou et al. 2022), i.e. closest to Q. cerris than to any of the other Quercus species sampled herein. Suber I may correspond to what has been considered the primary chloroplast lineage in Q. suber, and referred to as the “suber” chloroplast lineage, as opposed to the “ilex” chloroplast lineage (Lumaret et al. 2005; Simeone et al. 2018). Suber II appears either unresolved (MP and ML trees) or nested within Q. rotundifolia, without support (BEAST tree), and is separated from suber I, in the haplotype network, by six intermediate haplotypes of Q. rotundifolia and Q. cerris. This pattern raises questions on the origin and persistence of the two Q. suber chloroplast lineages, which could be explained by two biological scenarios. The first would be incomplete lineage sorting, meaning that Q. suber retained two ancestral chloroplast lineages within its distribution range. The second would be ancient introgression between Q. suber and Quercus sect. Ilex. Both these processes have been invoked to explain haplotype diversity in Quercus sect. Cerris (Simeone et al. 2018). Our analyses show an ancient origin of both Q. suber chloroplast lineages but also the presence of different haplogroups in Quercus sect. Ilex, as well as a lack of support for relationships between the two sections. These results are compatible with a scenario of incomplete lineage sorting, i.e. the persistence of ancestral chloroplast lineages within both Q. suber and Q. rotundifolia, but do not negate the possibility that haplotypes in suber II derive from introgression events between Q. suber and Q. rotundifolia/Q. ilex, as these two species were not sampled across their entire range, and a more complete sampling may have shown a pattern more indicative of hybridisation.
The recovery of the Q. ilex haplotype nested within suber II confirms the existence of genetic exchange between Q. ilex and Q. suber. Hybridisation between these two species has been widely described (Belahbib et al. 2001; López de Heredia et al. 2017; Lumaret et al. 2009) and Q. suber is considered to be the paternal donor in most cases (Belahbib et al. 2001; López de Heredia et al. 2020), although some authors do not recognise a directional hybridisation pattern in these two taxa (Burgarella et al. 2009; López de Heredia et al. 2017; Lumaret et al. 2009). The observed pattern suggests that the Q. ilex haplotype is derived from a Q. suber haplotype present in a diversified suber II group. However, both Q. ilex and Q. rotundifolia are not broadly sampled in our data set, and a putative introgression event where Q. ilex is the paternal donor is likely to be uncommon. The parsimony haplotype network supports the hypothesis of suber II originating through introgression, as the two Q. suber chloroplast groups are separated by intermediate haplotypes of Q. rotundifolia. However, haplotype network reconstruction is not robust to small changes in our data set. It was observed that the simple removal of Q. cerris and of parsimony-informative site 458, for example, results in a network showing the two suber haplotype groups connected, without intermediate haplotypes from other taxa (Online Resource 3).
Age estimates obtained under a relaxed clock recover the origin of Quercus at c. 55 Myr, in accordance with earlier analyses that placed the origin of the genus at 55–56 Myr (Hipp et al. 2020; Hubert et al. 2014). The option for a relaxed clock was considered appropriate given the inclusion of different Quercus species and the outgroup. Crown age estimates for the two suber haplotype groups and for the Q. suber/Q. cerris clade point to the late Oligocene/early Miocene, and are thus compatible with the dates proposed by Magri et al. (2007) for the diversification of Q. suber haplotypes. Our age estimates have a large margin of error, as indicated by the height posterior density (HPD) intervals, and must be considered as only an approximation. Nevertheless, these estimates suggest that the two haplotype lineages were present in Q. suber well before the Pleistocene, and therefore that putative introgression events with Q. ilex/Q. rotundifolia (Jiménez et al. 2004; Lumaret and Jabbour-Zahab 2009) originating suber II haplotypes would likely have occurred before glaciations associated with that epoch.
If the suber II haplogroup was indeed acquired through modern introgression between Q. suber and Q. ilex/Q. rotundifolia, the latter corresponding to seed-bearing donors, then all sampled suber II haplotypes in our trees would have to be derived from multiple unsampled Q. ilex or Q. rotundifolia haplotypes. Ancient introgression between Q. suber and an ancestral lineage in Quercus sect. Ilex, during the Miocene, could nevertheless be a valid possibility, assuming that both lineages were already well differentiated, although the Q. coccifera/Q. ilex haplotype split is estimated at c.9 Myr in our analysis (Hipp et al. 2020 estimated the species split at c.10 Myr), and is thus more recent than the suber II group. The hypothesis of ancient reticulations explaining the presence of different chloroplast lineages in Quercus sects. Cerris and Ilex has been postulated earlier (e.g. Simeone et al. 2016, 2018). These reticulations may have occurred as multiple independent events, and haplotypes would have become fixed due to genetic drift associated with demographic changes. However, our current sampling of Quercus sect. Ilex is insufficient to fully verify the hypothesis of ancient reticulation events originating the two distinct haplogroups.
An alternative explanation for the observed pattern would be incomplete sorting of chloroplast lineages in Q. suber. Manos et al. (1999) hypothesised the persistence of cpDNA polymorphisms through the diversification of Quercus, and highlighted the lack of a clear discriminating signal of Quercus plastid data at infrageneric level. Li et al. (2022) have reported long-term persistence of ancestral chloroplast lineages in East Asian oaks. Lumaret et al. (2005) also predicted the existence of two native Q. suber haplogroups, rather than a native and an introgressed lineage, and Simeone et al. (2009) postulated that sharing of ancestral cpDNA polymorphisms in sect. Cerris is highly probable.
Besides Q. suber, one of the hypothetical maternal donors under the introgression scenario, Q. rotundifolia, does not possess a single chloroplast lineage either. This pattern is consistent with the findings of Simeone et al. (2016) who reported a non-monophyly of plastomes in Quercus sect. Ilex, which was attributed to a combination of incomplete lineage sorting and putative introgression. Vitelli et al. (2017) also identified different lineages in Quercus sect. Ilex which followed a clear geographical structuring. Samples of Q. rotundifolia do not cluster together in our phylogenetic analyses, and indeed display a pattern that is compatible with the retention of different chloroplast lineages. For example, three Q. rotundifolia haplotypes (XII, XIII, XIV) form a supported clade with the Q. coccifera haplotypes (IV, V) in the BEAST tree (Fig. 3), and are readily identified in the haplotype network (Fig. 4). These haplotypes may correspond to what Simeone et al. (2016) named the “EuroMed” lineage of sect. Ilex plastomes.
Persistence of ancestral polymorphisms is a product of both rapid diversification and large effective population sizes (Pamilo and Nei 1988; Maddison and Knowles 2006). If ancestral Q. suber populations experienced rapid growth before a single chloroplast lineage could be fixed throughout the entire species range, then the two haplogroups could have persisted without major constraints for long periods. Diversification in Quercus species has been related to ecological opportunity due to a mid-Miocene temperature decrease (Graham 2011; Hipp et al. 2020), which may have promoted population expansion as well as speciation. Large effective population sizes throughout the Miocene may have enabled the retention of different chloroplast lineages up to the Pleistocene, when glaciation cycles caused range contractions in tree species north of the Mediterranean, that affected Q. suber (López de Heredia et al. 2007a; Vessella et al. 2015). Range contractions and decreased effective population sizes during glaciation periods would in theory have favoured the fixation of polymorphisms and chloroplast lineages. To explain the persistence of the two suber haplogroups, assuming they both have an ancestral origin, perhaps a vicariance process must be invoked. Vicariance could have resulted from contraction, during Pleistocene glacial maxima, into disconnected refugia in which either haplogroup, suber I or II, would become fixed. The geographical distribution of the suber II group may support the hypothesis of a separation of the two lineages, since Q. suber haplotypes in this group were only found in the Iberian Peninsula and northwestern Africa, whereas suber I is found throughout the distribution range of the species. Haplotypes of the suber II group may have become fixed in refugia located in the Iberian Peninsula and northwestern Africa (see Vessella et al. 2015), and post-glacial secondary contact between formerly isolated populations holding either suber I o II would then have allowed for the coexistence of the two haplogroups in these regions. Alternatively, new areas of suitable habitat for Q. suber may have become available around southern refugia, as hypothesised by Vesella et al. (2015), allowing for the maintenance of a large effective population and the presence of the two haplogroups. The current presence of the two lineages in the Iberian Peninsula and northwestern Africa but not in the remaining distribution range could be explained by the generally larger effective sizes of Q. suber populations in these regions, more recent colonisation of the northern and eastern parts of the species distribution range and low dispersal of suber I seeds across the Pyrenees, although a degree of adaptive leverage of either haplogroup cannot be excluded (see Pham et al. 2017 and López de Heredia et al. 2017). Regarding cpDNA diversity among regions, Algeria, Morocco, Spain, Italy and Portugal stand out as having large haplotype diversity and more than one private haplotype. Values of Tajima’s D statistic were generally negative, with statistical significance in France and Italy. This deviation from neutrality indicates recent population expansion. Like other Mediterranean tree species, Q.suber is known to have expanded northwards after contracting during the last glaciation period (López de Heredia et al. 2007a; Vessella et al. 2015), and signal of population expansion should therefore be detectable, particularly in the northern part of the Q. suber distribution range, where post-glacial colonisation is likely to be more recent.
Conclusions
We identified two cpDNA haplogroups within the distribution range of Q. suber, in agreement with earlier studies on Q. suber chloroplasts. One of these haplogroups occurs only in the Iberian Peninsula and northwestern Africa, while the other is present across the species range. Age estimates point to a Miocene diversification of both haplogroups, suggesting a scenario involving the retention of ancient chloroplast lineages or the occurrence of ancient introgression events between Q. suber and Q. sect. Ilex. Differential fixation of these chloroplast lineages in refugia and recent population expansion may explain their persistence through glaciation periods and their present-day geographical distribution. Our results highlight the complexity of chloroplast genealogies in Q. suber, Q. ilex and Q. rotundifolia, and suggest that obtaining more data, such as whole chloroplast sequences, may be required to fully understand the processes behind chloroplast diversity in these Mediterranean oak species.
Data availability
Aligned sequence data and scripts used for analyses are available at: https://github.com/CoBiG2/Qsuber_cpDNA.
References
Aronson J, Pereira JS, Pausas JG (eds) (2009) Cork oak woodlands on the edge: ecology, adaptive management, and restoration. Island Press, Washington
Bagnoli F, Tsuda Y, Fineschi S, Bruschi P, Magri D, Zhelev P, Paule L, Simeone MC, González-Martínez SC, Vendramin GG (2016) Combining molecular and fossil data to infer demographic history of Quercus cerris: insights on European eastern glacial refugia. J Biogeogr 43:679–690. https://doi.org/10.1111/jbi.12673
Belahbib N, Pemonge MH, Ouassou A, Sbay H, Kremer A, Petit RJ (2001) Frequent cytoplasmic exchanges between oak species that are not closely related: Quercus suber and Q. ilex in Morocco. Molec Ecol 10:2003–2012. https://doi.org/10.1046/j.0962-1083.2001.01330.x
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537. https://doi.org/10.1371/journal.pcbi.1003537
Burgarella C, Lorenzo Z, Jabbour-Zahab R, Lumaret R, Guichoux E, Petit RJ, Soto Á, Gil L (2009) Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex). Heredity 102:442–452. https://doi.org/10.1038/hdy.2009.8
Caudullo G, Welk E, San-Miguel-Ayanz J (2017) Chorological maps for the main European woody species. Data Brief 12:662–666. https://doi.org/10.1016/j.dib.2017.05.007
CBOL Plant Working Group (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106:12794–12797. https://doi.org/10.1073/pnas.0905845106
Costa J, Miguel C, Almeida H, Oliveira MM, Matos JA, Simões F, Veloso M, Pinto RC, Paulo OS, Batista D (2011) Genetic divergence in Cork Oak based on cpDNA sequence data. BMC Proc 5(Suppl 7):P13. https://doi.org/10.1186/1753-6561-5-S7-P13
Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002) Molecular hylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid and rbcl, atpB and matK DNA sequences. Amer J Bot 89:132–144
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Meth 9:772–772. https://doi.org/10.1038/nmeth.2109
de Dato GD, Teani A, Mattioni C, Aravanopoulos F, Avramidou EV, Stojnic S, Ganopoulos I, Belletti P, Ducci F (2020) Genetic analysis by nuSSR Markers of Silver Birch (Betula pendula Roth) populations in their Southern European distribution range. Frontiers Pl Sci 11:310. https://doi.org/10.3389/fpls.2020.00310
Denk T, Grimm GW, Manos PS, Deng M, Hipp AL (2017) An updated infrageneric classification of the oaks: review of previous taxonomic schemes and synthesis of evolutionary patterns. In: Gil-Pelegrín E, Peguero-Pina JJ, Sancho-Knapik D (eds), Exploring the functional diversity of genus Quercus L. Springer, Cham, pp 13–38. https://doi.org/10.1007/978-3-319-69099-5_2
Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24:774–786. https://doi.org/10.1111/j.1096-0031.2008.00217.x
Graham A (2011) The age and diversification of terrestrial New World ecosystems through Cretaceous and Cenozoic time. Amer J Bot 98:336–351. https://doi.org/10.3732/ajb.1000353
Hipp AL, Manos PS, Hahn M, Avishai M, Bodénès C, Cavender-Bares J, Crowl AA, Deng M, Denk T, Fitz-Gibbon S, Gailing O, González-Elizondo MS, González-Rodríguez A, Grimm GW, Jiang X-L, Kremer A, Lesur I, McVay JD, Plomion C, Rodríguez-Correa H, Schulze E-D, Simeone MC, Sork VL, Valencia-Avalos S (2020) Genomic landscape of the global oak phylogeny. New Phytol 226:1198–1212. https://doi.org/10.1111/nph.16162
Hochbach A, Linder HP, Röser M (2018) Nuclear genes, matK and the phylogeny of the Poales. Taxon 67:521–536. https://doi.org/10.12705/673.5
Hubert F, Grimm GW, Jousselin E, Berry V, Franc A, Kremer A (2014) Multiple nuclear genes stabilize the phylogenetic backbone of the genus Quercus. Syst Biodivers 12:405–423. https://doi.org/10.1080/14772000.2014.941037
Jiménez P, de Heredia UL, Collada C, Lorenzo Z, Gil L (2004) High variability of chloroplast DNA in three Mediterranean evergreen oaks indicates complex evolutionary history. Heredity 93:510–515. https://doi.org/10.1038/sj.hdy.6800551
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molec Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102:8369–8374. https://doi.org/10.1073/pnas.050312310
Li Y, Wang L, Zhang X, Kang H, Liu C, Mao L, Fang Y (2022) Extensive sharing of chloroplast haplotypes among East Asian Cerris oaks: the imprints of shared ancestral polymorphism and introgression. Ecol Evol 12:e9142. https://doi.org/10.1002/ece3.9142
López de Heredia U, Carrión JS, Jiménez P, Collada C, Gil L (2007a) Molecular and palaeoecological evidence for multiple glacial refugia for evergreen oaks on the Iberian Peninsula. J Biogeogr 34:1505–1517. https://doi.org/10.1111/j.1365-2699.2007.01715.x
López de Heredia U, Jiménez P, Collada C, Simeone MC, Bellarosa R, Schirone B, Cervera MT, Gil L (2007b) Multi-marker phylogeny of three evergreen oaks reveals vicariant patterns in the Western Mediterranean. Taxon 56:1209–1220. https://doi.org/10.2307/25065912
López-de-Heredia U, Jiménez P, Díaz-Fernández P, Gil L (2005) The Balearic Islands: a reservoir of cpDNA genetic variation for evergreen oaks. J Biogeogr 32:939–949. https://doi.org/10.1111/j.1365-2699.2004.01232.x
López de Heredia U, Mora-Márquez F, Goicoechea PG, Guillardín-Calvo L, Simeone MC, Soto Á (2020) ddRAD sequencing-based identification of genomic boundaries and permeability in Quercus ilex and Q. suber hybrids. Frontiers Pl Sci 11:564414. https://doi.org/10.3389/fpls.2020.564414
Lopez-Sanchez A, San Miguel A, Dirzo R, Roig S (2016) Scattered trees and livestock grazing as keystones organisms for sustainable use and conservation of Mediterranean dehesas. J Nat Conservation 33:58–67. https://doi.org/10.1016/j.jnc.2016.07.003
López de Heredia U, Vázquez FM, Soto Á (2017) The role of hybridization on the adaptive potential of Mediterranean sclerophyllous oaks: the case of the Quercus ilex x Q. suber complex. In: Gil-Pelegrín E, Peguero-Pina JJ, Sancho-Knapik D (eds), Oaks Physiological Ecology. Exploring the Functional Diversity of Genus Quercus L. Springer, Cham, pp 239–260
Lumaret R, Jabbour-Zahab R (2009) Ancient and current gene flow between two distantly related Mediterranean oak species, Quercus suber and Q. ilex. Ann Bot (Oxford) 104:725–736. https://doi.org/10.1093/aob/mcp149
Lumaret R, Tryphon-Dionnet M, Michaud H, Sanuy A, Ipotesi E, Born C, Mir C (2005) Phylogeographical variation of chloroplast DNA in cork oak (Quercus suber). Ann Bot (Oxford) 96:853–861. https://doi.org/10.1016/j.jnc.2016.07.003
Maddison WP, Knowles LL (2006) Inferring phylogeny despite incomplete lineage sorting. Syst Biol 55:21–30. https://doi.org/10.1080/10635150500354928
Magri D, Fineschi S, Bellarosa R, Buonamici A, Sebastiani F, Schirone B, Simeone MC, Vendramin GG (2007) The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Molec Ecol 16:5259–5266. https://doi.org/10.1111/j.1365-294X.2007.03587.x
Manos PS, Doyle JJ, Nixon KC (1999) Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). Molec Phylogen Evol 12:333–349. https://doi.org/10.1006/mpev.1999.0614
Mariac C, Scarcelli N, Pouzadou J, Barnaud A, Billot C, Faye A, Kougbeadjo A, Maillol V, Martin G, Sabot F, Santoni S, Vigouroux Y, Couvreur TL (2014) Cost-effective enrichment hybridization capture of chloroplast genomes at deep multiplexing levels for population genetics and phylogeography studies. Molec Ecol Resources 14:1103–1113. https://doi.org/10.1111/1755-0998.12258
Miller MA, Pfeiffer W, Schwartz T (2011) The CIPRES science gateway: a community resource for phylogenetic analyses. In: Proceedings of the 2011 TeraGrid Conference: extreme digital discovery, pp 1–8. https://doi.org/10.1145/2016741.2016785
Modesto IS, Miguel C, Pina-Martins F, Glushkova M, Veloso M, Paulo OS, Batista D (2014) Identifying signatures of natural selection in cork oak (Quercus suber L.) genes through SNP analysis. Tree Genet Genomes 10:1645–1660. https://doi.org/10.1007/s11295-014-0786-1
Nishizawa T, Watano Y (2000) Primer pairs suitable for PCR-SSCP analysis of chloroplast DNA in angiosperms. J Phytogeogr Taxon 48:63–66
Pamilo P, Nei M (1988) Relationships between gene trees and species trees. Molec Biol Evol 5:568–583. https://doi.org/10.1093/oxfordjournals.molbev.a040517
Paradis E (2010) pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26:419–420
Petit RJ, Csaikl UM, Bordács S, Burg K, Coart E, Cottrell J, van Dam B, Deans JD, Dumolin-Lapègue S, Fineschi S, Finkeldey R, Gillies A, Glaz I, Goicoechea PG, Jensen JS, König AO, Lowe AJ, Madsen SF, Mátyás G, Munro RC, Olalde M, Pemonge M-H, Popescu F, Slade D, Tabbener H, Taurchini D, de Vries SGM, Ziegenhagen B, Kremer A (2002) Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. Forest Ecol Managem 156:5–26. https://doi.org/10.1016/S0378-1127(01)00645-4
Petit RJ, Vendramin GG (2007) Plant phylogeography based on organelle genes: an introduction. In: Weiss S, Ferrand N (eds) Phylogeography of Southern European Refugia. Springer, Dordrecht, pp 23–97. https://doi.org/10.1007/1-4020-4904-8_2
Pham KK, Hipp AL, Manos PS, Cronn RC (2017) A time and a place for everything: phylogenetic history and geography as joint predictors of oak plastome phylogeny. Genome 60:720–732. https://doi.org/10.1139/gen-2016-0191
Pina-Martins F, Baptista J, Pappas G Jr, Paulo OS (2019) New insights into adaptation and population structure of cork oak using genotyping by sequencing. Global Change Biol 25:337–350. https://doi.org/10.1111/gcb.14497
Piredda R, Simeone MC, Attimonelli M, Bellarosa R, Schirone B (2011) Prospects of barcoding the Italian wild dendroflora: oaks reveal severe limitations to tracking species identity. Molec Ecol Resources 11:72–83. https://doi.org/10.1111/j.1755-0998.2010.02900.x
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. Available at: http://www.R-project.org/
Ramírez-Valiente JA, Sánchez-Gómez D, Aranda I, Valladares F (2010) Phenotypic plasticity and local adaptation in leaf ecophysiological traits of 13 contrasting cork oak populations under different water availabilities. Tree Physiol 30:618–627. https://doi.org/10.1093/treephys/tpq013
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Molec Biol Evol 19:2092–2100. https://doi.org/10.1093/oxfordjournals.molbev.a004034
Rolo V, Moreno G (2019) Shrub encroachment and climate change increase the exposure to drought of Mediterranean wood-pastures. Sci Total Environm 660:550–558. https://doi.org/10.1016/j.scitotenv.2019.01.029
Simeone MC, Cardoni S, Piredda R, Imperatori F, Avishai M, Grimm GW, Denk T (2018) Comparative systematics and phylogeography of Quercus Section Cerris in western Eurasia: inferences from plastid and nuclear DNA variation. PeerJ 6:e5793. https://doi.org/10.7717/peerj.5793
Simeone MC, Grimm GW, Papini A, Vessella F, Cardoni S, Tordoni E, Piredda R, Franc A, Denk T (2016) Plastome data reveal multiple geographic origins of Quercus Group Ilex. PeerJ 4:e1897. https://doi.org/10.7717/peerj.1897
Simeone MC, Papini A, Vessella F, Bellarosa R, Spada F, Schirone B (2009) Multiple genome relationships and a complex biogeographic history in the eastern range of Quercus suber L.(Fagaceae) implied by nuclear and chloroplast DNA variation. Caryologia 62:236–252. https://doi.org/10.1080/00087114.2004.10589689
Simeone MC, Piredda R, Papini A, Vessella F, Schirone B (2013) Application of plastid and nuclear markers to DNA barcoding of Euro-Mediterranean oaks (Quercus, Fagaceae): problems, prospects and phylogenetic implications. Bot J Linn Soc 172:478–499. https://doi.org/10.1111/boj.12059
Soto A, Lorenzo Z, Gil L (2007) Differences in fine-scale genetic structure and dispersal in Quercus ilex L. and Q. suber L.: consequences for regeneration of Mediterranean open woods. Heredity 99:601–607. https://doi.org/10.1038/sj.hdy.6801007
Sousa F, Bertrand YJ, Nylinder S, Oxelman B, Eriksson JS, Pfeil BE (2014) Phylogenetic properties of 50 nuclear loci in Medicago (Leguminosae) generated using multiplexed sequence capture and next-generation sequencing. PLoS ONE 9:e109704. https://doi.org/10.1371/journal.pone.0109704
Sousa F, Costa J, Ribeiro C, Varandas M, Pina-Martins F, Simões F, Matos J, Glushkova M, Miguel C, Veloso MM, Oliveira M, Ricardo CP, Batista D, Paulo OS (2022) Population structure in Quercus suber L. revealed by nuclear microsatellite markers. PeerJ 10:e13565. https://doi.org/10.7717/peerj.13565
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Staudt M, Mir C, Joffre R, Rambal S, Bonin A, Landais D, Lumaret R (2004) Isoprenoid emissions of Quercus spp (Q. suber and Q. ilex) in mixed stands contrasting in interspecific genetic introgression. New Phytol 163:573–584. https://doi.org/10.1111/j.1469-8137.2004.01140.x
Taberlet P, Fumagalli L, Wust-Saucy AG, Cossons J-F (1998) Comparative phylogeography and postglacial colonization routes in Europe. Molec Ecol 7:453–464. https://doi.org/10.1046/j.1365-294x.1998.00289.x
Tajima F (1989) The effect of change in population size on DNA polymorphism. Genetics 123:597–601. https://doi.org/10.1093/genetics/123.3.597
Vanhove M, Pina-Martins F, Coelho AC, Branquinho C, Costa A, Batista D, Príncipe A, Sousa P, Henriques A, Marques I, Belkadi B, Knowles LL, Paulo OS (2021) Using gradient Forest to predict climate response and adaptation in Cork oak. J Evol Biol 34:910–923. https://doi.org/10.1111/jeb.13765
Vessella F, López-Tirado J, Simeone MC, Schirone B, Hidalgo PJ (2017) A tree species range in the face of climate change: cork oak as a study case for the Mediterranean biome. Eur J Forest Res 136:555–569. https://doi.org/10.1007/s10342-017-1055-2
Vessella F, Simeone MC, Schirone B (2015) Quercus suber range dynamics by ecological niche modelling: from the Last Interglacial to present time. Quatern Sci Rev 119:85–93. https://doi.org/10.1016/j.quascirev.2015.04.018
Vila-Viçosa C, Gonçalves J, Honrado J, Lomba Â, Almeida RS, Vázquez FM, Garcia C (2020) Late Quaternary range shifts of marcescent oaks unveil the dynamics of a major biogeographic transition in southern Europe. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-78576-9
Vitelli M, Vessella F, Cardoni S, Pollegioni P, Denk T, Grimm GW, Simeone MC (2017) Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae). Tree Genet Genomes 13:1–17. https://doi.org/10.1007/s11295-016-1086-8
Zhou BF, Yuan S, Crowl AA, Liang YY, Shi Y, Chen XY, An QQ, Kang M, Manos PS, Wang B (2022) Phylogenomic analyses highlight innovation and introgression in the continental radiations of Fagaceae across the Northern Hemisphere. Nat Commun 13:1–14. https://doi.org/10.1038/s41467-022-28917-1
Acknowledgements
This work was supported by project ‘Keep Pace: Selection of trees keeping pace with fast environmental changes, a science-based approach for sustainable XXI century Oak forests’, co-funded by the EU ERDF funds, within the Portugal 2020 Partnership Agreement and Programa Operacional Regional Algarve, and by national funds through Fundação para a Ciência e Tecnologia (FCT) Portugal (ALG-01-0145-FEDER-029263 / PTDC/ASP/SIL/29263/2017). Additional support was received through grants PTDC/AGR-GPL/104966/2008 and SOBREIRO/0036/2009, and through the research units GREEN-IT, Bioresources for Sustainability (UIDB/04551/2020) and cE3c (UIDB/00329/2020).
Funding
Open access funding provided by FCT|FCCN (b-on).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Filipe de Sousa, Joana Costa, Isabel Marques, Dora Batista, Octávio S. Paulo. The first draft of the manuscript was written by Filipe de Sousa, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Handling Editor: Michał Ronikier.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. List of samples showing the corresponding sampling sites and haplotypes.
Online Resource 2. Maximum-parsimony consensus tree of combined chloroplast data.
Online Resource 3. Haplotype parsimony network after site and sample exclusion.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sousa, F., Viegas, M.B., Costa, J. et al. Haplotype diversity patterns in Quercus suber (Fagaceae) inferred from cpDNA sequence data. Plant Syst Evol 309, 42 (2023). https://doi.org/10.1007/s00606-023-01879-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-023-01879-7