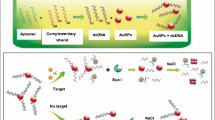
Abstract
GOLD SELEX, a novel SELEX approach has been developed that obviates the need for target immobilization for aptamer development. The approach purely relies on the affinity of the aptamers towards its target, to get detached from the gold nanoparticle (GNP) surface (weak attraction) after binding with its target. Thus, only the completely detached aptamers are selected for the next round of SELEX. This, in-process, also addresses the issue of residual binding and thus improves the sensitivity of the developed aptamers. As a proof of concept for establishing the utility of the approach for small molecules, we have developed aptamers against dichlorvos (DV), a pesticide in just 8 rounds. Using these aptamer candidates, we have developed an aptamer-NanoZyme (GNP having peroxidase mimic activity) based colorimetric assay. The developed aptamer displayed high affinity (Kd in sub micromolar range) and selectivity for DV. The developed assay could detect as low as 15 μM DV. The best-performing aptamer was also able to work in real samples like river water and commercial apple juice. The GOLD SELEX approach developed in this study, we believe, can act as a template for future SELEX strategy development and can replace the conventional SELEX strategy.
Graphical abstract









Similar content being viewed by others
References
Sharma TK, Bruno JG, Dhiman A (2017) ABCs of DNA aptamer and related assay development. Biotechnol Adv 35:275–301
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: Chemi-SELEX. Science (80- ) 249:505–510. https://doi.org/10.1038/346818a0
Wu J, Zhu Y, Xue F, Mei Z, Yao L, Wang X, Zheng L, Liu J, Liu G, Peng C, Chen W (2014) Recent trends in SELEX technique and its application to food safety monitoring. Microchim Acta 181:479–491. https://doi.org/10.1007/s00604-013-1156-7
Chatterjee B, Jyoti Das S, Anand A, Kumar Sharma T (2020) Nanozymes and aptamer-based biosensing. Mater Sci Energy Technol 3:127–135. https://doi.org/10.1016/j.mset.2019.08.007
Ciancio DR, Vargas MR, Thiel WH et al (2018) Aptamers as diagnostic tools in cancer. Pharmaceuticals 11:1–23. https://doi.org/10.3390/ph11030086
Ye H, Duan N, Wu S, Tan G, Gu H, Li J, Wang H, Wang Z (2017) Orientation selection of broad-spectrum aptamers against lipopolysaccharides based on capture-SELEX by using magnetic nanoparticles. Microchim Acta 184:4235–4242. https://doi.org/10.1007/s00604-017-2453-3
Tian F, Zhou J, Jiao B, He Y (2019) A nanozyme-based cascade colorimetric aptasensor for amplified detection of ochratoxin A. Nanoscale 11:9547–9555. https://doi.org/10.1039/c9nr02872b
Dhiman A, Kalra P, Bansal V et al (2017) Aptamer-based point-of-care diagnostic platforms. Sensors Actuators B Chem 246:535–553. https://doi.org/10.1016/j.snb.2017.02.060
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822. https://doi.org/10.1016/0021-9797(80)90501-9
Stoltenburg R, Nikolaus N, Strehlitz B (2012) Capture-SELEX: selection of DNA aptamers for aminoglycoside antibiotics. J Anal Methods Chem 1:1–14. https://doi.org/10.1155/2012/415697
Yu H, Yang W, Alkhamis O, Canoura J, Yang KA, Xiao Y (2018) In vitro isolation of small-molecule-binding aptamers with intrinsic dye-displacement functionality. Nucleic Acids Res 46:e43–e43. https://doi.org/10.1093/nar/gky026
Pfeiffer F, Mayer G (2016) Selection and biosensor application of aptamers for small molecules. Front Chem 4:1–21. https://doi.org/10.3389/fchem.2016.00025
Ruscito A, DeRosa MC (2016) Small-molecule binding aptamers: selection strategies, characterization, and applications. Front Chem 4:1–14. https://doi.org/10.3389/fchem.2016.00014
Qiao N, Li J, Wu X, Diao D, Zhao J, Li J, Ren X, Ding X, Shangguan D, Lou X (2019) Speeding up in vitro discovery of structure-switching speeding up in vitro discovery of structure-switching aptamers via magnetic cross-linking precipitation. Anal Chem 91:13383–13389. https://doi.org/10.1021/acs.analchem.9b00081
Nutiu R, Li Y (2005) In vitro selection of structure-switching signaling aptamers. Angew Chem Int Ed 44:1061–1065. https://doi.org/10.1002/anie.200461848
Oh SS, Plakos K, Lou X, Xiao Y, Soh HT (2010) In vitro selection of structure-switching, self-reporting aptamers. Proc Natl Acad Sci U S A 107:14053–14058. https://doi.org/10.1073/pnas.1009172107
Sher M, Zhuang R, Demirci U, Asghar W (2017) Paper-based analytical devices for clinical diagnosis: recent advances in the fabrication techniques and sensing mechanisms. Expert Rev Mol Diagn 17:351–366. https://doi.org/10.1080/14737159.2017.1285228
Kaur H, Bruno JG, Kumar A, Sharma TK (2018) Aptamers in the therapeutics and diagnostics pipelines. Theranostics 8:4016–4032. https://doi.org/10.7150/thno.25958
Cheeveewattanagul N, Morales-Narváez E, Hassan ARHA, Bergua JF, Surareungchai W, Somasundrum M, Merkoçi A (2017) Straightforward Immunosensing platform based on graphene oxide-decorated nanopaper: a highly sensitive and fast biosensing approach. Adv Funct Mater 27:1–8. https://doi.org/10.1002/adfm.201702741
Li W, Luo Y, Gao T, Yang L, Wang J, Pei R (2019) In vitro selection of DNA aptamers for a small-molecule porphyrin by gold nanoparticle-based SELEX. J Mol Evol 87:231–239. https://doi.org/10.1007/s00239-019-09905-4
Li H, Rothberg L (2004) Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Natl Acad Sci U S A 101:14036–14039. https://doi.org/10.1073/pnas.0406115101
Sharma TK, Ramanathan R, Weerathunge P, Mohammadtaheri M, Daima HK, Shukla R, Bansal V (2014) Aptamer-mediated “turn-off/turn-on” nanozyme activity of gold nanoparticles for kanamycin detection. Chem Commun 50:15856–15859. https://doi.org/10.1039/c4cc07275h
Chatterjee B, Kalyani N, Das S, et al (2019) Nano-realm for point-of-care (POC) bacterial diagnostics. In: Methods in microbiology, 1st ed. Elsevier Ltd., pp 19–42
Abnous K, Danesh NM, Ramezani M et al (2018) A colorimetric gold nanoparticle aggregation assay for malathion based on target-induced hairpin structure assembly of complementary strands of aptamer. Microchim Acta 185:5–11
Zhao W, Chiuman W, Brook MA, Li Y (2007) Simple and rapid colorimetric biosensors based on DNA aptamer and noncrosslinking gold nanoparticle aggregation. ChemBioChem 8:727–731. https://doi.org/10.1002/cbic.200700014
Weerathunge P, Ramanathan R, Shukla R, Sharma TK, Bansal V (2014) Aptamer-controlled reversible inhibition of gold nanozyme activity for pesticide sensing. Anal Chem 86:11937–11941. https://doi.org/10.1021/ac5028726
Alsager OA, Alotaibi KM, Alswieleh AM, Alyamani BJ (2018) Colorimetric Aptasensor of vitamin D3: a novel approach to eliminate residual adhesion between Aptamers and Gold nanoparticles. Sci Rep 8:12947. https://doi.org/10.1038/s41598-018-31221-y
Liu J, Bai W, Niu S, et al (2014) Highly sensitive colorimetric detection of 17β-estradiol using split DNA aptamers immobilized on unmodified gold nanoparticles. Sci Rep 4
Lee BH, Nguyen VT, Gu MB (2017) Highly sensitive detection of 25-HydroxyvitaminD3 by using a target-induced displacement of aptamer. Biosens Bioelectron 88:174–180. https://doi.org/10.1016/j.bios.2016.08.011
Okoroiwu HU, Iwara IA (2018) Dichlorvos toxicity: a public health perspective. Interdiscip Toxicol 11:129–137. https://doi.org/10.2478/intox-2018-0009
Wei M, Feng S (2017) Amperometric determination of organophosphate pesticides using a acetylcholinesterase based biosensor made from nitrogen-doped porous carbon deposited on a boron-doped diamond electrode. Microchim Acta 184:3461–3468. https://doi.org/10.1007/s00604-017-2380-3
Hertz-Picciotto I, Sass JB, Engel S, Bennett DH, Bradman A, Eskenazi B, Lanphear B, Whyatt R (2018) Organophosphate exposures during pregnancy and child neurodevelopment: recommendations for essential policy reforms. PLoS Med 15:1–15. https://doi.org/10.1371/journal.pmed.1002671
Chemical concoction: pesticides in veggies and fruits harm Hyderabad children | Hyderabad News - Times of India. https://timesofindia.indiatimes.com/city/hyderabad/chemical-concoction-pesticides-in-veggies-and-fruits-harm-hyderabad-children/articleshow/65126763.cms. Accessed 21 Jan 2020
India needs a comprehensive approach to prevent pesticide related suicides. https://www.downtoearth.org.in/blog/agriculture/india-needs-a-comprehensive-approach-to-prevent-pesticide-related-suicides-61587. Accessed 21 Jan 2020
Das R, Dhiman A, Kapil A, Bansal V, Sharma TK (2019) Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal Bioanal Chem 411:1229–1238
Kalra P, Subodh Kumar Mishra SK, Amit Kumar HKP et al (2018) G-Quadruplex-forming DNA aptamers inhibit the DNA-binding function of HupB and Mycobacterium tuberculosis entry into host cells. Mol Ther - Nucleic Acids 13:99–109. https://doi.org/10.1016/j.omtn.2018.08.011
Dhiman A, Haldar S, Mishra SK, Sharma N, Bansal A, Ahmad Y, Kumar A, Sharma TK, Tyagi JS (2018) Generation and application of DNA aptamers against HspX for accurate diagnosis of tuberculous meningitis. Tuberculosis 112:27–36. https://doi.org/10.1016/j.tube.2018.07.004
Khan E, Biswas S, Mishra SK, Mishra R, Samanta S, Mishra A, Tawani A, Kumar A (2019) Rationally designed small molecules targeting toxic CAG repeat RNA that causes Huntington’s disease (HD) and spinocerebellar ataxia (SCAs). Biochimie 163:21–32. https://doi.org/10.1016/j.biochi.2019.05.001
Khan E, Tawani A, Mishra SK, Verma AK, Upadhyay A, Kumar M, Sandhir R, Mishra A, Kumar A (2018) Myricetin reduces toxic level of CAG repeats RNA in Huntington’s disease (HD) and Spino cerebellar Ataxia (SCAs). ACS Chem Biol 13:180–188. https://doi.org/10.1021/acschembio.7b00699
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Zumrut HE, Batool S, Argyropoulos KV, Williams N, Azad R, Mallikaratchy PR (2019) Integrating ligand-receptor interactions and in vitro evolution for streamlined discovery of artificial nucleic acid ligands. Mol Ther - Nucleic Acids 17:150–163. https://doi.org/10.1016/j.omtn.2019.05.015
Das R, Chaterjee B, Kapil A, Sharma TK (2020) Aptamer-NanoZyme mediated sensing platform for the rapid detection of Escherichia coli in fruit juice. Sens Bio-Sens Res 27:100313. https://doi.org/10.1016/j.sbsr.2019.100313
Jv Y, Li B, Cao R (2010) Positively-charged gold nanoparticles as peroxidiase mimic and their application in hydrogen peroxide and glucose detection. Chem Commun 46:8017–8019. https://doi.org/10.1039/c0cc02698k
Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA (2011) NUPACK: analysis and design of nucleic acid systems. J Comput Chem 32:170–173. https://doi.org/10.1002/jcc.21596
Chen X, Wang J, Jiang G, Zu G, Liu M, Zhou L, Pei R (2016) The development of a light-up red-emitting fluorescent probe based on a G-quadruplex specific cyanine dye. RSC Adv 6:70117–70123. https://doi.org/10.1039/c6ra11152a
Das A, Bhadra K, Kumar GS (2011) Targeting RNA by small molecules: comparative structural and thermodynamic aspects of aristololactam-β-D-glucoside and daunomycin binding to tRNA phe. PLoS One 6:e23186. https://doi.org/10.1371/journal.pone.0023186
Buurma NJ, Haq I (2007) Advances in the analysis of isothermal titration calorimetry data for ligand-DNA interactions. Methods 42:162–172. https://doi.org/10.1016/j.ymeth.2007.01.010
Calderone CT, Williams DH (2001) An enthalpic component in cooperativity: the relationship between enthalpy, entropy, and noncovalent structure in weak associations. J Am Chem Soc 123:6262–6267
Craddock HA, Huang D, Turner PC, Quirós-Alcalá L, Payne-Sturges DC (2019) Trends in neonicotinoid pesticide residues in food and water in the United States, 1999-2015. Environ Health 18:1–16. https://doi.org/10.1186/s12940-018-0441-7
Zhang WJ, Bin JF, Ou JF (2011) Global pesticide consumption and pollution: with China as a focus. Proc Int Acad Ecol Environ Sci 1:125–144. https://doi.org/10.0000/issn-2220-8860-piaees-2011-v1-0012
Hyland C, Bradman A, Gerona R, Patton S, Zakharevich I, Gunier RB, Klein K (2019) Organic diet intervention significantly reduces urinary pesticide levels in U.S. children and adults. Environ Res 171:568–575. https://doi.org/10.1016/j.envres.2019.01.024
Thuy PT, van Geluwe S, Nguyen VA, van der Bruggen B (2012) Current pesticide practices and environmental issues in Vietnam: management challenges for sustainable use of pesticides for tropical crops in (South-East) Asia to avoid environmental pollution. J Mater Cycles Waste Manag 14:379–387. https://doi.org/10.1007/s10163-012-0081-x
Most samples exceeding fipronil limit from the Netherlands. https://www.foodnavigator.com/Article/2018/05/07/Most-samples-exceeding-fipronil-limit-from-the-Netherlands. Accessed 23 Jan 2020
Taiwan food chiefs hatch new egg safety plan to crack down on fipronil fears. https://www.foodnavigator-asia.com/Article/2019/05/29/Taiwan-food-chiefs-hatch-new-egg-safety-plan-to-crack-down-on-fipronil-fears. Accessed 23 Jan 2020
Kaur H, Chatterjee B, Bruno JG, Sharma TK (2019) Defining target product profiles (TPPs) for aptamer-based diagnostics. In: Advances in biochemical engineering/biotechnology. Springer, Berlin, pp 1–12
Hermann T, Patel DJ (2000) Adaptive recognition by nucleic acid aptamers. Science (80- ) 287:820–825. https://doi.org/10.1126/science.287.5454.820
Linnet K, Kondratovich M (2004) Partly nonparametric approach for determining the limit of detection. Clin Chem 50:732–740. https://doi.org/10.1373/clinchem.2003.029983
Johansson MA, Hellenäs KE (2003) Immunobiosensor analysis of clenbuterol in bovine hair. Food Agric Immunol 15:197–205. https://doi.org/10.1080/09540100400011058
D’Souza SL, Pati RK, Kailasa SK (2014) Ascorbic acid functionalized gold nanoparticles as a probe for colorimetric and visual read-out determination of dichlorvos in environmental samples. Anal Methods 6:9007–9014. https://doi.org/10.1039/c4ay01004c
Shrivastava A, Gupta V (2011) Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chronicles Young Sci 2:21. https://doi.org/10.4103/2229-5186.79345
Jalalian SH, Karimabadi N, Ramezani M, Abnous K, Taghdisi SM (2018) Electrochemical and optical aptamer-based sensors for detection of tetracyclines. Trends Food Sci Technol 73:45–57. https://doi.org/10.1016/j.tifs.2018.01.009
Liu M, Khan A, Wang Z, Liu Y, Yang G, Deng Y, He N (2019) Aptasensors for pesticide detection. Biosens Bioelectron 130:174–184. https://doi.org/10.1016/j.bios.2019.01.006
Majdinasab M, Mitsubayashi K, Marty JL (2019) Optical and electrochemical sensors and biosensors for the detection of quinolones. Trends Biotechnol 37:898–915. https://doi.org/10.1016/j.tibtech.2019.01.004
McKeague M, McConnell EM, Cruz-Toledo J et al (2015) Analysis of in vitro aptamer selection parameters. J Mol Evol 81:150–161. https://doi.org/10.1007/s00239-015-9708-6
Musheev MU, Krylov SN (2006) Selection of aptamers by systematic evolution of ligands by exponential enrichment: addressing the polymerase chain reaction issue. Anal Chim Acta 564:91–96. https://doi.org/10.1016/j.aca.2005.09.069
Shao K, Ding W, Wang F, Li H, Ma D, Wang H (2011) Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS One 6:1–7. https://doi.org/10.1371/journal.pone.0024910
Tolle F, Wilke J, Wengel J, Mayer G (2014) By-product formation in repetitive PCR amplification of DNA libraries during SELEX. PLoS One 9:1–12. https://doi.org/10.1371/journal.pone.0114693
Wilson C, Nix J, Szostak J (1998) Functional requirements for specific ligand recognition by a biotin- binding RNA pseudoknot. Biochemistry 37:14410–14419. https://doi.org/10.1021/bi981371j
Mannironi C, Di Nardo A, Fruscoloni P, Tocchini-Valentini GP (1997) In vitro selection of dopamine RNA ligands. Biochemistry 36:9726–9734. https://doi.org/10.1021/bi9700633
Nelson EM, Rothberg LJ (2011) Kinetics and mechanism of single-stranded DNA adsorption onto citrate-stabilized gold nanoparticles in colloidal solution. Langmuir 27:1770–1777. https://doi.org/10.1021/la102613f
Demers LM, Östblom M, Zhang H, Jang NH, Liedberg B, Mirkin CA (2002) Thermal desorption behavior and binding properties of DNA bases and nucleosides on gold. J Am Chem Soc 124:11248–11249. https://doi.org/10.1021/ja0265355
Zhang X, Servos MR, Liu J (2012) Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir 28:3896–3902. https://doi.org/10.1021/la205036p
Kwon YS, Ahmad Raston NH, Gu MB (2014) An ultra-sensitive colorimetric detection of tetracyclines using the shortest aptamer with highly enhanced affinity. Chem Commun 50:40–42. https://doi.org/10.1039/c3cc47108j
Tian Y, Wang Y, Sheng Z, Li T, Li X (2016) A colorimetric detection method of pesticide acetamiprid by fine-tuning aptamer length. Anal Biochem 513:87–92. https://doi.org/10.1016/j.ab.2016.09.004
Alsager OA, Kumar S, Zhu B, Travas-Sejdic J, McNatty KP, Hodgkiss JM (2015) Ultrasensitive colorimetric detection of 17-estradiol: the effect of shortening DNA aptamer sequences. Anal Chem 87:4201–4209. https://doi.org/10.1021/acs.analchem.5b00335
Kalra P, Dhiman A, Cho WC, Bruno JG, Sharma TK (2018) Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front Mol Biosci 5:1–16. https://doi.org/10.3389/fmolb.2018.00041
Gao S, Zheng X, Jiao B, Wang L (2016) Post-SELEX optimization of aptamers. Anal Bioanal Chem 408:4567–4573. https://doi.org/10.1007/s00216-016-9556-2
Acknowledgments
The aptamer sequences mentioned in the manuscript is deposited to Indian Patent Office (Patent No. 202011004692).
Funding
This work was supported through the THSTI Core Grant and Innovative Young Biotechnologist Award (IYBA), grant number (BT/010/IYBA/2016/10) to TKS. BC and AA were supported by the Biotechnology Industrial Research Assistance Council (BIRAC), Govt. of India in the form of post-doctoral and Technical Assistant position respectively through BIRAC-PACE grant to TKS. NK was recipient of Department of Science and Technology (DST) National Post-Doctoral Fellowship (NPDF). The excellent technical assistance of Mr. Rajkumar Dwivedi is duly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
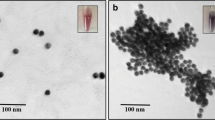
Supplementary material includes chemical structures of the pesticides, percentage base composition of sequences, multiple sequence alignment of the sequences, phylogenetic dendrogram, Sequences and length of the selected aptamers, TEM images of the GNPs, UV-Vis spectra, peroxidase activity graph, LOD and LOQ calculation. It is available in the online version of this article at https://doi.org/10.1007/s12274-***-****-* (automatically inserted by the publisher). (DOCX 4.18 mb)
Rights and permissions
About this article
Cite this article
Chatterjee, B., Kalyani, N., Anand, A. et al. GOLD SELEX: a novel SELEX approach for the development of high-affinity aptamers against small molecules without residual activity. Microchim Acta 187, 618 (2020). https://doi.org/10.1007/s00604-020-04577-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-04577-0