Abstract
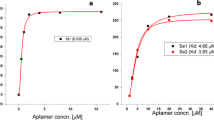
The authors describe a new kind of selection method (referred to as orientation selection) for improved screening for broad-spectrum lipopolysaccharides (LPSs) using unlabeled ssDNA aptamers. The method is based on the capture-SELEX technique using magnetic nanoparticles. LPS from Salmonella enterica serotype typhimurium was chosen as an exemplary target. Once the ssDNA library is preconcentrated to a certain degree, two Gram-negative bacteria were used as orientation molecules in the subsequent selection process. Using this strategy, one optimal aptamer ('EA7') was captured that has a high affinity (Kd = 102 ± 17 nM) for LPS and it was also confirmed that it can recognize three other bacterial LPSs. It is presumed that EA7 binds to the lipid A region of LPS. When using carboxyfluorescein labeled EA7, the observed fluorescence intensity and concentrations of four types of LPSs in drinking water are linearly correlated. The lower detection limit of the LPS is 3 ng·mL−1. Compared to multi-target mixed selection and conventional SELEX methods, the new orientation selection strategy produces results that are less uncertain.

An improved orientation selection strategy in capture-SELEX procedures was developed to screen broad-spectrum aptamers against lipopolysaccharides by using magnetic nanoparticles. Two Gram-negative bacteria were used as orientation molecules after library preconcentrated to a certain degree.





Similar content being viewed by others
References
Erridge C, Bennett-Guerrero E, Poxton IR (2002) Structure and function of lipopolysaccharides. Microbes Infect 4(8):15
Raetz CR, Whitfield C (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi:10.1146/annurev.biochem.71.110601.135414
Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D (2016) Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14(6):337–345. doi:10.1038/nrmicro.2016.25
Wang X, Zhang C, Shi F, Hu X (2010) Purification and characterization of lipopolysaccharides. Subcell Biochem 53:27–51. doi:10.1007/978-90-481-9078-2_2
Xu W, Tian J, Shao X, Zhu L, Huang K, Luo Y (2016) A rapid and visual aptasensor for lipopolysaccharides detection based on the bulb-like triplex turn-on switch coupled with HCR-HRP nanostructures. Biosens Bioelectron. doi:10.1016/j.bios.2016.10.012
Mack L, Brill B, Delis N, Groner B (2014) Endotoxin depletion of recombinant protein preparations through their preferential binding to histidine tags. Anal Biochem 466:83–88. doi:10.1016/j.ab.2014.08.020
Sanghavi BJ, Wolfbeis OS, Hirsch T, Swami NS (2015) Nanomaterial-based electrochemical sensing of neurological drugs and neurotransmitters. Microchim Acta 182:1–41. doi:10.1007/s00604-014-1308-4
Rajendran M, Ellington AD (2008) Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal Bioanal Chem 390(4):1067–1075. doi:10.1007/s00216-007-1735-8
Wu C, Han D, Chen T, Peng L, Zhu G, You M, Qiu L, Sefah K, Zhang X, Tan W (2013) Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J Am Chem Soc 135(49):18644–18650. doi:10.1021/ja4094617
Wu J, Zhu Y, Xue F, Mei Z, Yao L, Wang X, Zheng L, Liu J, Liu G, Peng C, Chen W (2014) Recent trends in SELEX technique and its application to food safety monitoring. Microchim Acta 181(5–6):479–491. doi:10.1007/s00604-013-1156-7
Wu S, Wang Y, Duan N, Ma H, Wang Z (2015) Colorimetric Aptasensor based on enzyme for the detection of vibrio parahemolyticus. J Agric Food Chem 63(35):7849–7854. doi:10.1021/acs.jafc.5b03224
Zhu C, Zhang G, Huang Y, Yan J, Chen A (2016) Aptamer based ultrasensitive determination of the β-adrenergic agonist ractopamine using PicoGreen as a fluorescent DNA probe. Microchim Acta 184(2):439–444. doi:10.1007/s00604-016-2032-z
Abnous K, Danesh NM, Alibolandi M, Ramezani M, Taghdisi SM (2017) Amperometric aptasensor for ochratoxin a based on the use of a gold electrode modified with aptamer, complementary DNA, SWCNTs and the redox marker methylene blue. Microchim Acta 184(4):1151–1159. doi:10.1007/s00604-017-2113-7
Bunka DH, Stockley PG (2006) Aptamers come of age - at last. Nat Rev Microbiol 4(8):588–596. doi:10.1038/nrmicro1458
Zhou J, Rossi J (2016) Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. doi:10.1038/nrd.2016.199
Aquino-Jarquin G, Toscano-Garibay JD (2011) RNA aptamer evolution: two decades of SELEction. Int J Mol Sci 12(12):9155–9171. doi:10.3390/ijms12129155
Wang J, Gong Q, Maheshwari N, Eisenstein M, Arcila ML, Kosik KS, Soh HT (2014) Particle display: a quantitative screening method for generating high-affinity aptamers. Angew Chem Int Ed Engl 53(19):4796–4801. doi:10.1002/anie.201309334
Fabian Spill ZBW, Shemirani AI, Ho N, Desai D, Zaman MH (2016) Controlling uncertainty in aptamer selection. PNAS 113(43):12076–12081
Chen X, Huang Y, Duan N, Wu S, Xia Y, Ma X, Zhu C, Jiang Y, Ding Z, Wang Z (2014) Selection and characterization of single stranded DNA aptamers recognizing fumonisin B1. Microchim Acta 181(11–12):1317–1324. doi:10.1007/s00604-014-1260-3
Stoltenburg R, Nikolaus N, Strehlitz B (2012) Capture-SELEX: selection of DNA aptamers for aminoglycoside antibiotics. J Anal Methods Chem 2012:415697. doi:10.1155/2012/415697
Wu Y, Zhan S, Wang L, Zhou P (2014) Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles. Analyst 139(6):1550–1561. doi:10.1039/c3an02117c
Paniel N, Istamboulie G, Triki A, Lozano C, Barthelmebs L, Noguer T (2017) Selection of DNA aptamers against penicillin G using capture-SELEX for the development of an impedimetric sensor. Talanta 162:232–240. doi:10.1016/j.talanta.2016.09.058
Spiga FM, Maietta P, Guiducci C (2015) More DNA-aptamers for small drugs: a capture-SELEX coupled with surface Plasmon resonance and high-throughput sequencing. ACS Comb Sci 17(5):326–333. doi:10.1021/acscombsci.5b00023
Duan N, Gong W, Wu S, Wang Z (2017) Selection and application of ssDNA aptamers against Clenbuterol hydrochloride based on ssDNA library immobilized SELEX. J Agric Food Chem 65(8):1771–1777. doi:10.1021/acs.jafc.6b04951
Wang L, Liu X, Zhang Q, Zhang C, Liu Y, Tu K, Tu J (2012) Selection of DNA aptamers that bind to four organophosphorus pesticides. Biotechnol Lett 34(5):869–874. doi:10.1007/s10529-012-0850-6
Wen AQ, Yang QW, Li JC, Lv FL, Zhong Q, Chen CY (2009) A novel lipopolysaccharide-antagonizing aptamer protects mice against endotoxemia. Biochem Biophys Res Commun 382(1):140–144. doi:10.1016/j.bbrc.2009.02.152
Bruno JG, Carrillo MP, Phillips T (2008) In vitro antibacterial effects of antilipopolysaccharide DNA aptamer-C1qrs complexes. Folia Microbiol 53(4):8
Ying G, Zhu F, Yi Y, Chen J, Mei J, Zhang Y, Chen S (2015) Selecting DNA aptamers for endotoxin separation. Biotechnol Lett 37(8):1601–1605. doi:10.1007/s10529-015-1839-8
Ding JL, Gan ST, Ho B (2009) Single-stranded DNA oligoaptamers: molecular recognition and LPS antagonism are length- and secondary structure-dependent. J Innate Immun 1(1):46–58. doi:10.1159/000145542
Wu S, Duan N, Wang Z, Wang H (2011) Aptamer-functionalized magnetic nanoparticle-based bioassay for the detection of ochratoxin a using upconversion nanoparticles as labels. Analyst 136(11):2306–2314. doi:10.1039/c0an00735h
Wang L, Ye H, Sang H-Q, Wang D-D (2016) Aptamer-based fluorescence assay for detection of Isocarbophos and Profenofos. Chin J Anal Chem 44(5):799–803. doi:10.1016/s1872-2040(16)60933-7
Yang KA, Barbu M, Halim M, Pallavi P, Kim B, Kolpashchikov DM, Pecic S, Taylor S, Worgall TS, Stojanovic MN (2014) Recognition and sensing of low-epitope targets via ternary complexes with oligonucleotides and synthetic receptors. Nat Chem 6(11):1003–1008. doi:10.1038/nchem.2058
Acknowledgments
This work was supported by National Natural Science Fund of China [21375049], the special fund for innovation in inspection and detection of food and drug of GDFDA [2015ZX07], Key Research and Development Program of Jiangsu Province [BE2017623, BE2016306], the natural science foundation of Jiangsu Province [BK20140155], China Postdoctoral Science Foundation [2016 T90430], and the Technology Research and Development Program of Suzhou [SYN201513].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOCX 614 kb)
Rights and permissions
About this article
Cite this article
Ye, H., Duan, N., Wu, S. et al. Orientation selection of broad-spectrum aptamers against lipopolysaccharides based on capture-SELEX by using magnetic nanoparticles. Microchim Acta 184, 4235–4242 (2017). https://doi.org/10.1007/s00604-017-2453-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2453-3