Abstract
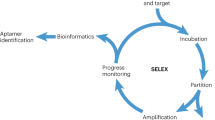
Aptamers are functional single-stranded DNA or RNA oligonucleotides, selected in vitro by SELEX (Systematic Evolution of Ligands by Exponential Enrichment), which can fold into stable unique three-dimensional structures that bind their target ligands with high affinity and specificity. Although aptamers show a number of favorable advantages such as better stability and easier modification when compared with the properties of antibodies, only a handful of aptamers have entered clinical trials and only one, pegaptanib, has received US Food and Drug Administration approval for clinical use. The main reasons that limit the practical application of aptamers are insufficient nuclease stability, bioavailability, thermal stability, or even affinity. Some aptamers obtained from modified libraries show better properties; however, polymerase amplification of nucleic acids containing non-natural bases is currently a primary drawback of the SELEX process. This review focuses on several post-SELEX optimization strategies of aptamers identified in recent years. We describe four common methods in detail: truncation, chemical modification, bivalent or multivalent aptamer construction, and mutagenesis. We believe that these optimization strategies should improve one or more specific properties of aptamers, and the type of feature(s) selected for improvement will be dependent on the application purpose.

Similar content being viewed by others
References
Shigdar S, Macdonald J, O’Connor M, Wang T, Xiang DX, Al Shamaileh H, et al. Aptamers as theranostic agents: modifications, serum stability and functionalisation. Sensors (Basel). 2013;13(10):13624–37. doi:10.3390/s131013624.
Wang RE, Wu H, Niu Y, Cai J. Improving the stability of aptamers by chemical modification. Curr Med Chem. 2011;18(27):4126–38.
Radom F, Jurek PM, Mazurek MP, Otlewski J, Jelen F. Aptamers: molecules of great potential. Biotechnol Adv. 2013;31(8):1260–74. doi:10.1016/j.biotechadv.2013.04.007.
Shigdar S, Macdonald J, O’Connor M, Wang T, Xiang D, Al Shamaileh H, et al. Aptamers as theranostic agents: modifications, serum stability and functionalisation. Sensors. 2013;13(10):13624–37. doi:10.3390/s131013624.
Griffin LC, Tidmarsh GF, Bock LC, Toole JJ, Leung LL. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood. 1993;81(12):3271–6.
Lapa SA, Chudinov AV, Timofeev EN. The toolbox for modified aptamers. Mol Biotechnol. 2015. doi:10.1007/s12033-015-9907-9.
McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012;2012:748913. doi:10.1155/2012/748913.
Eaton BE, Gold L, Hicke BJ, Janjic N, Jucker FM, Sebesta DP, et al. Post-SELEX combinatorial optimization of aptamers. Bioorg Med Chem. 1997;5(6):1087–96.
Djordjevic M. SELEX experiments: new prospects, applications and data analysis in inferring regulatory pathways. Biomol Eng. 2007;24(2):179–89. doi:10.1016/j.bioeng.2007.03.001.
Kato Y, Minakawa N, Komatsu Y, Kamiya H, Ogawa N, Harashima H, et al. New NTP analogs: the synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res. 2005;33(9):2942–51. doi:10.1093/nar/gki578.
Pasternak A, Hernandez FJ, Rasmussen LM, Vester B, Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011;39(3):1155–64. doi:10.1093/nar/gkq823.
Cowperthwaite MC, Ellington AD. Bioinformatic analysis of the contribution of primer sequences to aptamer structures. J Mol Evol. 2008;67(1):95–102. doi:10.1007/s00239-008-9130-4.
Zheng X, Hu B, Gao SX, Liu DJ, Sun MJ, Jiao BH, et al. A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon. 2015;101:41–7. doi:10.1016/j.toxicon.2015.04.017.
Gao S, Hu B, Zheng X, Cao Y, Liu D, Sun M, et al. Gonyautoxin 1/4 aptamers with high-affinity and high-specificity: from efficient selection to aptasensor application. Biosens Bioelectron. 2016;79:938–44. doi:10.1016/j.bios.2016.01.032.
Nadal P, Svobodova M, Mairal T, O'Sullivan CK. Probing high-affinity 11-mer DNA aptamer against Lup an 1 (beta-conglutin). Anal Bioanal Chem. 2013;405(29):9343–9. doi:10.1007/s00216-013-7385-0.
Sung HJ, Choi S, Lee JW, Ok CY, Bae YS, Kim YH, et al. Inhibition of human neutrophil activity by an RNA aptamer bound to interleukin-8. Biomaterials. 2014;35(1):578–89. doi:10.1016/j.biomaterials.2013.09.107.
Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W. Optimization and modifications of aptamers selected from live cancer cell lines. Chembiochem. 2007;8(6):603–6. doi:10.1002/cbic.200600532.
Kong HY, Byun J. Nucleic acid aptamers: new methods for selection, stabilization, and application in biomedical science. Biomol Ther. 2013;21(6):423–34. doi:10.4062/biomolther.2013.085.
Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi:10.1038/nature06783.
Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi:10.1126/science.1178178.
Lee KY, Kang H, Ryu SH, Lee DS, Lee JH, Kim S. Bioimaging of nucleolin aptamer-containing 5-(N-benzylcarboxyamide)-2′-deoxyuridine more capable of specific binding to targets in cancer cells. J Biomed Biotechnol. 2010. doi:10.1155/2010/168306.
Yamamoto T, Nakatani M, Narukawa K, Obika S. Antisense drug discovery and development. Future Med Chem. 2011;3(3):339–65. doi:10.4155/fmc.11.2.
King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37(47):16489–93. doi:10.1021/bi981780f.
Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–83. doi:10.1146/annurev.med.56.062904.144915.
Reinemann C, Strehlitz B. Aptamer-modified nanoparticles and their use in cancer diagnostics and treatment. Swiss Med Wkly. 2014;144:w13908. doi:10.4414/smw.2014.13908.
Wang J, Li G. Aptamers against cell surface receptors: selection, modification and application. Curr Med Chem. 2011;18(27):4107–16.
Dhakal S, Yu Z, Konik R, Cui Y, Koirala D, Mao H. G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys J. 2012;102(11):2575–84. doi:10.1016/j.bpj.2012.04.024.
Tatarinova O, Tsvetkov V, Basmanov D, Barinov N, Smirnov I, Timofeev E, et al. Comparison of the ‘chemical’ and ‘structural’ approaches to the optimization of the thrombin-binding aptamer. PloS One. 2014;9(2):e89383. doi:10.1371/journal.pone.0089383.
Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, et al. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32(19):5757–65. doi:10.1093/nar/gkh862.
Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, et al. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011;39(6):2458–69. doi:10.1093/nar/gkq996.
Kim Y, Cao Z, Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc Natl Acad Sci U S A. 2008;105(15):5664–9. doi:10.1073/pnas.0711803105.
Nonaka Y, Yoshida W, Abe K, Ferri S, Schulze H, Bachmann TT, et al. Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system. Anal Chem. 2013;85(2):1132–7. doi:10.1021/ac303023d.
Muller J, Freitag D, Mayer G, Potzsch B. Anticoagulant characteristics of HD1-22, a bivalent aptamer that specifically inhibits thrombin and prothrombinase. J Thromb Haemost. 2008;6(12):2105–12. doi:10.1111/j.1538-7836.2008.03162.x.
Musumeci D, Montesarchio D. Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer. Pharmacol Ther. 2012;136(2):202–15. doi:10.1016/j.pharmthera.2012.07.011.
Kim Y, Dennis DM, Morey T, Yang L, Tan WH. Engineering dendritic aptamer assemblies as superior inhibitors of protein function. Chem-Asian J. 2010;5(1):56–9. doi:10.1002/asia.200900421.
Zhao X, Lis JT, Shi H. A systematic study of the features critical for designing a high avidity multivalent aptamer. Nucleic Acid Ther. 2013;23(3):238–42. doi:10.1089/nat.2012.0410.
Bullock TL, Sherlin LD, Perona JJ. Tertiary core rearrangements in a tight binding transfer RNA aptamer. Nat Struct Biol. 2000;7(6):497–504. doi:10.1038/75910.
Cho JS, Lee SW. Sequence and structural features of RNA aptamer against myasthenic autoantibodies. Oligonucleotides. 2009;19(3):273–80. doi:10.1089/oli.2009.0201.
Moore MD, Cookson J, Coventry VK, Sproat B, Rabe L, Cranston RD, et al. Protection of HIV neutralizing aptamers against rectal and vaginal nucleases: implications for RNA-based therapeutics. J Biol Chem. 2011;286(4):2526–35. doi:10.1074/jbc.M110.178426.
Hoshika S, Minakawa N, Matsuda A. Synthesis and physical and physiological properties of 4′-thioRNA: application to post-modification of RNA aptamer toward NF-kappaB. Nucleic Acids Res. 2004;32(13):3815–25. doi:10.1093/nar/gkh705.
Kaur H, Li JJ, Bay BH, Yung LY. Investigating the antiproliferative activity of high affinity DNA aptamer on cancer cells. PLoS One. 2013;8(1):e50964. doi:10.1371/journal.pone.0050964.
Pedersen EB, Nielsen JT, Nielsen C, Filichev VV. Enhanced anti-HIV-1 activity of G-quadruplexes comprising locked nucleic acids and intercalating nucleic acids. Nucleic Acids Res. 2011;39(6):2470–81. doi:10.1093/nar/gkq1133.
Forster C, Zydek M, Rothkegel M, Wu Z, Gallin C, Gessner R, et al. Properties of an LNA-modified ricin RNA aptamer. Biochem Biophys Res Commun. 2012;419(1):60–5. doi:10.1016/j.bbrc.2012.01.127.
Kolb G, Reigadas S, Boiziau C, van Aerschot A, Arzumanov A, Gait MJ, et al. Hexitol nucleic acid-containing aptamers are efficient ligands of HIV-1 TAR RNA. Biochemistry. 2005;44(8):2926–33. doi:10.1021/bi048393s.
Reinemann C, Strehlitz B. Aptamer-modified nanoparticles and their use in cancer diagnostics and treatment. Swiss Med Wkly. 2014;144:w13908. doi:10.4414/smw.2014.13908.
Shiang YC, Huang CC, Wang TH, Chien CW, Chang HT. Aptamer-conjugated nanoparticles efficiently control the activity of thrombin. Adv Funct Mater. 2010;20(18):3175–82. doi:10.1002/adfm.201000642.
Acknowledgments
The authors thank Professor Jiao and Professor Wang for guidance and review of the manuscript and the National High-Tech Research and Development Program of China for funding this work (2013AA092904).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Shunxiang Gao and Xin Zheng contributed equally to this work.
Rights and permissions
About this article
Cite this article
Gao, S., Zheng, X., Jiao, B. et al. Post-SELEX optimization of aptamers. Anal Bioanal Chem 408, 4567–4573 (2016). https://doi.org/10.1007/s00216-016-9556-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9556-2