Abstract
Cynomolgus macaques are widely used as a primate model for human diseases associated with an immunological process. Because there are individual differences in immune responsiveness, which are controlled by the polymorphic nature of the major histocompatibility (MHC) locus, it is important to reveal the diversity of MHC in the model animal. In this study, we analyzed 26 cynomolgus macaques from five families for MHC class I genes. We identified 32 Mafa-A, 46 Mafa-B, 6 Mafa-I, and 3 Mafa-AG alleles in which 14, 20, 3, and 3 alleles were novel. There were 23 MHC class I haplotypes and each haplotype was composed of one to three Mafa-A alleles and one to five Mafa-B alleles. Family studies revealed that there were two haplotypes which contained two Mafa-A1 alleles. These observations demonstrated further the complexity of MHC class I locus in the Old World monkey.
Similar content being viewed by others
Introduction
Non-human primates are widely used for immunological research because their immune system is similar to that of humans. In particular, the Old World monkeys such as cynomolgus macaques (crab-eating macaques, Macaca fascicularis) became a useful model for human infectious diseases including acquired immunodeficiency syndrome (AIDS) (Wiseman et al. 2007), severe acute respiratory syndrome (Lawler et al. 2006), and influenza (Kobasa et al. 2007) as well as in the transplantation field (Wiseman and O’Connor 2007). In the AIDS research, cynomolgus and rhesus macaques are important animal models for the development of vaccines against human immunodeficiency virus (HIV) or studies for susceptibility to HIV infection and/or development of AIDS (Matano et al. 2004; Loffredo et al. 2008; Tsukamoto et al. 2008; Burwitz et al. 2009; Mee et al. 2009; Aarnink et al. 2011a). To fully evaluate the results of immunological experiments in the macaque models, it is essential to characterize the genetic diversity of immune-related molecules which may control the individual differences in the immune response against foreign antigens and/or pathogens.
The major histocompatibility complex (MHC) is well known to control the immune-responsiveness to foreign antigens. There are two classes of MHC molecules: one is the MHC class I molecule presenting peptides of intracellular origin to CD8+ T cell and the other is the MHC class II molecule binding extracellular-derived antigenic peptides for presenting to CD4+ T cell. It has been reported that the complexity of MHC genes in the rhesus and cynomolgus macaques is higher than that in humans (Kulski et al. 2004; Watanabe et al. 2006; Gibbs et al. 2007; Otting et al. 2007, 2008; Doxiadis et al. 2011). For example, MHC class I configurations in macaques are usually composed of one copy of highly transcribed major MHC-A1gene (Mamu-A1or Mafa-A1) and several other minor MHC-A genes (Mamu-A2~A7 or Mafa-A2~A6) in addition to several MHC-B genes (Mamu-B or Mafa-B) (Watanabe et al. 2006; Otting et al. 2007, 2008, 2009; Naruse et al. 2010; Doxiadis et al. 2011), whereas each one copy of MHC-A and -B genes (HLA-A and -B) can be found in human MHC class I locus. In addition, other MHC loci showing lower expression levels, i.e., HLA-B-like gene (Mamu-I or Mafa-I) and HLA-G-like non-classical gene (Mamu-AG or Mafa-AG) have been identified (Slukvin et al. 2000; Urvater et al. 2000). The extent of genetic diversity is different, in part, depending on the geographic areas, as we have previously reported for MHC class I genes in rhesus macaque (Naruse et al. 2010). As for the cynomolgus macaques, MHC class I allelic diversity was reported for Indonesian (Pendley et al. 2008; Wu et al. 2008; Kita et al. 2009; Otting et al. 2009), Malaysian (Otting et al. 2009; Aarnink et al. 2011b), Mauritian (Budde et al. 2010), Vietnamese (Wu et al. 2008; Kita et al. 2009), and Philippino (Campbell et al. 2009; Kita et al. 2009) macaques, but information about the MHC class I haplotype remains insufficient.
In the present study, we have analyzed MHC class I loci in cynomolgus macaques originated from Indonesia, Malaysia, and the Philippines to obtain information on haplotype configuration. We report here further the complex nature of MHC class I loci in the Old World monkey, i.e., the presence of unique haplotypes carrying two Mafa-A1 genes.
Materials and methods
Animals
A total of 26 cynomolgus macaques from five families were the subjects. Each family was composed of one or two males with one or two females and their offspring. They were maintained in the breeding colonies in Tsukuba Primate Research Center, National Institute of Biomedical Innovation, Japan. The founders of the colonies were captured in Indonesia, Malaysia, and the Philippines. All care including blood sampling of animals were in accordance with the Guidelines for the Care and Use of Laboratory Animals published by the National Institute of Health (NIH Publication 85–23, revised 1985) and were subjected to prior approval by the local animal protection authority.
Sequencing analysis of cDNAs from Mafa class I genes
Total cellular RNA was extracted from whole blood by using RNAeasy (QIAGEN, Gmbh, Germany). Oligo(dT)-primed cDNA was synthesized using Transcriptor reverse transcriptase (Roche, Mannheim, Germany) according to the manufacturer’s recommendations. Full-length cDNAs for Mafa class I genes were amplified by polymerase chain reaction (PCR), as described previously (Tanaka-Takahashi et al. 2007; Naruse et al. 2010), by using locus-specific primer pairs as reported by Karl et al. (2008). Genomic gene and cDNA for Mafa-A2 gene were analyzed according to the method described by Wu et al. (2008). The primers used in this study are listed in Table 1. To estimate the expression level of Mafa-A alleles, we also used an additional primer pair: MafaF (5′-TACGTGGACGACACGCAGTT) and MafaR (5′-GGTGGGTCACATGTGTCTTG). PCR was done under the condition of initial denaturation at 98°C for 10 s, 25 cycles of 98°C for 1 s, 64°C for 5 s, and 72°C for 20 s, followed by an additional extension at 72°C for 1 min, using Phusion Flash DNA polymerase (Finzymes, Espoo, Finland). The PCR products were cloned into pSTBlue-1 Perfectly Blunt vector (Novagen, WI, USA) according to the manufacturer’s instructions and were transformed to NovaBlue Giga Singles™ competent cells (Merck Biosciences Japan, Tokyo, Japan). A total of 30 to 90 independent cDNA clones were obtained from each macaque for each locus and were sequenced on both strands by BigDye Terminator cycling system in an ABI 3730 automated sequence analyzer (Applied Biosystems, CA, USA).
Data analyses and nomenclature for Mafa class I allele
Nucleotide sequences of cDNA clones were aligned using the Genetyx software package (version 8.0, Genetyx Corp., Japan). When a cDNA sequence, which was represented by at least three clones, was independently obtained from at least two animals or repeatedly obtained from at least two independently prepared cDNAs from single animals, we considered it a real allele, not an artifact, and the sequences were submitted to the DNA Data Bank of Japan (DDBJ) database and to the Immuno Polymorphism Database for non-human primate MHC (http://www.ebi.ac.uk/ipd/mhc/sumit.html; Robinson et al. 2003) to obtain official nomenclature for the novel alleles of Mafa-A and Mafa-B genes. Neighbor-joining trees were constructed with Kimura’s two-parameter method for a phylogenetic analysis of Mafa-A sequences spanning exons 2, 3, and a part of exon 4 obtained in this study by using the Genetyx software. Bootstrap values were based on 5,000 replications.
Results
Identification of Mafa class I alleles in cynomolgus macaques
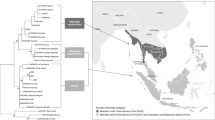
We determined the nucleotide sequences of cDNA clones for Mafa-A and -B loci in 26 cynomolgus macaques from one family of Indonesian origin (six haplotypes), two families of Malaysian origin (eight haplotypes), and two families of Philippino origin (nine haplotypes) (Fig. 1). When the observed alleles were segregated in the family or when at least three clones with identical sequences were observed from two independent PCR for an individual, the nucleotide sequences were considered to be real and not artifacts. As shown in Table 2, 32 Mafa-A, 46 Mafa-B, 6 Mafa-I, and 3 Mafa-AG sequences were obtained in this study. Among them, 14 (43.7%), 20 (43.5%), 3 (50.0%), and 3 (100%) were novel alleles of Mafa-A, Mafa-B, Mafa-I, and Mafa-AG loci, respectively (Table 2).
Pedigree of cynomolgus macaques. The pedigrees of macaques analyzed in this study are shown. Founders were originated from Indonesia (a), Malaysia (b, c), and Philippines (d, e). Open square, open circle, and open triangles indicate father, mother, and offspring, respectively. The ID of each subject is noted in the symbol. Mafa class I haplotypes determined in this study are indicated under the subjects
The Mafa-A alleles found in this study are listed in Table 3, where 21 alleles were from the major Mafa-A1 locus, while the remaining 11 alleles were from the minor Mafa-A loci, 3 from Mafa-A2, 3 from Mafa-A3, 2 from Mafa-A4, and 1 from Mafa-A6 alleles (Table 3). The major Mafa-A1alleles were defined by the sequence similarity to the known Mafa-A1 alleles to be given official nomenclatures by IPD, except for Mafa-A1*008:03-like allele, and we confirmed that the frequencies of cDNA clones for Mafa-A1 alleles were over 10% in each macaque. Similarly, alleles of minor Mafa-A genes, Mafa-A2, -A3, -A4, and -A6 were defined by sequence similarity to the known alleles. They, except for two novel Mafa-A2 alleles, were also given official names by IPD. On the other hand, a total of 46 Mafa-B alleles (Table 4) as well as 6 Mafa-I and 3 Mafa-AG alleles (Table 5) were identified. It was found that 2 out of 21 (9.5%) Mafa-A1a alleles and 12 out of 46 (26.1%) Mafa-B alleles had identical sequences to Mamu-A1 and Mamu-B alleles, respectively, implying a genetic admixture of cynomolgus macaques with rhesus macaques during the evolution (Otting et al. 2007; Bonhomme et al. 2009; Otting et al. 2009). Because we determined the nucleotide sequences only for exons 2, 3, and 4, two novel Mafa-AG alleles and three novel Mafa-I alleles were not given official names. As for the geographic distribution of Mafa class I alleles, there was no overlapping of Mafa-A alleles originated from different regions (Table 3), while there were a few Mafa-B and Mafa-I alleles commonly observed in macaques from different regions (Tables 4 and 5, respectively). When we looked into the presence of novel alleles in the geographic distribution, most of the novel alleles were obtained from Malaysian macaques, while almost all of the alleles found in Philippino macaques were not novel (Table 2).
Mafa class I haplotypes identified in the family study
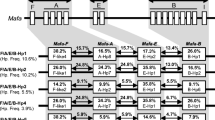
We could identify the Mafa-A and Mafa-B alleles composing 23 different haplotypes from the segregation studies (Table 6). It was found that one to three expressing Mafa-A alleles and one to five expressing Mafa-B alleles consisted of Mafa class I haplotype, similar to the Mamu class I haplotypes in rhesus macaques (Naruse et al. 2010). Of particular interest was that there were two haplotypes, “e” (Malaysian founder P03) and “v” (Philippino founder M05), carrying two different Mafa-A1 genes (Fig. 1; Table 6). Because previous studies have demonstrated that there is usually only one Mafa-A1 allele on a chromosome (Otting et al. 2007), while the presence of two Mamu-A1 alleles on the same haplotype was suggested in rhesus macaques (Naruse et al. 2010; Doxiadis et al. 2011), we performed further analyses.
The family studies showed that the Mafa-A1alleles consisting of haplotype “e”, Mafa-A1*001:01 andMafa-A1*032:05, or haplotype “v”, Mafa-A1*074:02 and Mafa-A1*093:01, did not carry accompanying minor Mafa-A genes (Table 6). When we constructed a phylogenetic tree of Mafa-A alleles identified in this study (Fig. 2), it was found that Mafa-A1*001:01 was mapped in the neighbor of Mafa-A3gene, raising a possibility that one of the two alleles on the same chromosome might be a minor Mafa-A allele and not the major Mafa-A1 allele. To test the possibility, we investigate the expression level of Mafa-A alleles composing of haplotypes “e” and “v”. For this purpose, other primer pairs were designed within the sequences completely shared by these alleles to amplify the Mafa-A cDNAs to avoid a possibility of affecting the efficacy of PCR by mismatches with the primer sequences. The cloning and sequencing analysis revealed that both Mafa-A1*001:01 and Mafa-A1*032:05 on the haplotype “e” were observed at similar frequencies among the cDNA clones of Mafa-A alleles in P03and C008 (Fig. 1): 29.7% and 33.3% in P03 and 22.5% and 17.5% in C008, respectively. Similarly, frequencies of haplotype “v” alleles, Mafa-A1*074:02 and Mafa-A1*093:01, in cDNA clones were 59.5% and 40.5%, respectively, in M05, while those in C010 were 23.3% and 26.7% and 31.4% and 17.1% in C011, respectively. The frequencies of cDNA clones varied in different individuals presumably due to the allelic competition with the alleles of another haplotype in each individual (Fig. 1), but they were much higher than the frequencies of the minor Mafa-A allele (Mafa-A3*13:03) clones: 3.3% and 2.9% in C010 and C011, respectively. These observations indicated that two Mafa-A alleles were considered to be major Mafa-A1 alleles in both haplotypes “e” and “v”.
Phylogenetic tree of Mafa-A alleles. A phylogenetic tree of the Mafa-A alleles detected in this study was constructed by using the neighbor-joining method with a bootstrap value of 5,000 replications. Values more than 50% are indicated as percentages. Novel alleles were underlined. Mafa-A1 alleles consisting of haplotype “e” are boxed, while the stippled boxes represent the alleles on haplotype “v”. Alleles of minor Mafa-A genes, Mafa-A2, A3, A4, and A6, are also indicated
Discussion
Native cynomolgus macaques are widespread throughout the islands of Southeast Asia into mainland Asia. They are mainly found in Indonesia, Malaysia, and the Philippines, then Burma, India, Vietnam, Cambodia, Laos, and Thailand (Lang 2006). It was suggested that the founding population of Mauritian macaques was introduced from Indonesia (Pendley et al. 2008; Campbell et al. 2009). More than 40% of Mafa class I alleles observed in this study were novel, even though there have been many reports on the analysis of Mafa class I genes, demonstrating that the diversity of MHC in the cynomolgus macaques still needs to be investigated. When we considered the origin of founders, 73.7% (28/38) were novel in alleles found in Malaysian macaques, while only 15.6% (5/32) were novel alleles in Philippino macaques (Table 2). The geographic distribution of novel alleles may be due to the fact that the Malaysian macaques had not been extensively analyzed before (Otting et al. 2007; Pendley et al. 2008; Kita et al. 2009). In the present study, B*089:01:02 was found in individuals among Indonesian, Malaysian, and Philippino macaques in different Mafa-B haplotypes (Table 6). Likewise, B*137:03 was found in Indonesian and Malaysian macaques (Table 4). In addition, shared alleles among the cynomolgus macaques, rhesus macaques, and pig-tailed macaques (Macaca nemestrina) were noted (Tables 3, 4, and 5). These observations indicated that the diversity of MHC class I genes is similar not only in the cynomolgus macaque population but also among the Old World monkeys, suggesting that the MHC class I polymorphisms might be generated before the divergence of Old World monkeys and/or there were admixtures of the Old World monkeys.
In this study, we determined the haplotype structure of Mafa class I locus by family studies and a total of 23 haplotypes were identified. Among them, haplotypes “i” and “w” carried identical Mafa-B alleles but different Mafa-A alleles (Table 6), suggesting that there were haplotypes originated by a recombination between the Mafa-A and Mafa-B loci. We showed that the Mafa class I haplotypes were usually composed of one to three Mafa-A alleles and one to five Mafa-B alleles, similar to the Mamu class I haplotypes, of which usually one MHC-A1 gene and a few (one to three) MHC-B genes were highly transcribed (Otting et al. 2007, 2008; Naruse et al. 2010; Doxiadis et al. 2011). As for the MHC-A locus in the cynomolgus macaques, highly transcribed Mafa-A1gene and other minor Mafa-A genes, such as Mafa-A2, -A3, -A4, and -A6 could be detected. It was reported that 87% of cynomolgus macaques had at least one Mafa-A2 alleles (Wu et al. 2008). However, only 3 out of 23 (13.0%) haplotypes carried a Mafa- A2 allele in this study (Table 6). We could not exclude a possibility that the strategy of our study might not be sufficient to detect the Mafa-A genes with low expression and/or the alleles with mismatches at the primer site, based on the number of clones within a PCR sample. Such a possibility is unlikely because we used the primer pairs which could cover the known Mafa-A2 alleles, although there might be novel Mafa-A2 alleles having different sequences at the primer binding sites. Therefore, we might underestimate the complexity of Mafa class I alleles in this study. High-throughput pyrosequencing methods may be a useful strategy to avoid the possibility of missing alleles, as described by several investigators (Wiseman et al. 2009; Budde et al. 2010; Aarnink et al. 2011b). In addition, because it was reported that the cell surface expression of Mamu class I molecule was varied depending on the locus and allelic structure (Rosner et al. 2010), locus- and allele-dependent expression of Mafa class I molecule at the cell surface will be required.
The most important finding in this study was that we demonstrated evidence for the presence of haplotypes carrying two major MHC-A1 genes on the same chromosome from the family studies and additional cloning studies. Interestingly, we and others have reported similar phenomena in rhesus macaques (Naruse et al. 2010; Doxiadis et al. 2011). In addition, several haplotypes carried multiple major Mafa-B1 alleles (Table 6), similar to the Mamu-B1 locus (Otting et al. 2008; Doxiadis et al. 2011). The raison d’etre of multiple major MHC class I genes/alleles on the same chromosome may be that they play an immunological role as the “double lock strategy” (Doxiadis et al. 2011) in which the double MHC-A1 alleles of high transcription level might be favorable to present peptide to CD8+ T cells. However, there is another unique haplotype which carries no MHC-A1allele in cynomolgus macaques (Otting et al. 2007) and maybe in rhesus macaques (Doxiadis et al. 2011). These observations suggested that the diversity of MHC in the Old World monkey is far more complicated than in humans.
In summary, we investigated 26 cynomolgus macaques from five families for the diversity of MHC class I alleles and haplotypes. A total of 87 alleles were identified, of which 40 were novel. There were 23 different haplotypes, and two of them carried two MHC-A1 genes, demonstrating further the complexity of MHC class I locus in the Old World monkey.
References
Aarnink A, Dereuddre-Bosquet N, Vaslin B, Le Grand R, Winterton P, Apoil PA, Blancher A (2011a) Influence of the MHC genotype on the progression of experimental SIV infection in the Mauritian cynomolgus macaque. Immunogenetics 63:267–274
Aarnink A, Apoil PA, Takahashi I, Osada N, Blancher (2011b) Characterization of MHC class I transcripts of a Malaysian cynomolgus macaque by high-throughput pyrosequencing and EST libraries. Immunogenetics. doi:10.1007/s00251-011-0550-8
Bonhomme M, Cuartero S, Blancher A, Crouau-Roy B (2009) Assessing natural introgression in 2 biomedical model species, the rhesus macaque (Macaca mulatta) and the long-tailed macaque (Macaca fascicularis). J Hered 100:158–169
Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, O’connor DH (2010) Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 62:773–780
Burwitz BJ, Pendley CJ, Greene JM, Detmer AM, Lhost JJ, Karl JA, Piaskowski SM, Rudersdorf RA, Wallace LT, Bimber BN, Loffredo JT, Cox DG, Bardet W, Hildebrand W, Wiseman RW, O’Connor SL, O’Connor DH (2009) Mauritian cynomolgus macaques share two exceptionally common major histocompatibility complex class I alleles that restrict simian immunodeficiency virus-specific CD8+ T cells. J Virol 83:6011–6019
Campbell KJ, Detmer AM, Karl JA, Wiseman RW, Blasky AJ, Hughes AL, Bimber BN, O’Connor SL, O’Connor DH (2009) Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics 61:177–187
Doxiadis GG, de Groot N, Otting N, Blokhuis JH, Bontrop RE (2011) Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics 63:73–83
Gibbs RA, Rogers J, Katze MG et al (2007) Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234
Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, Read DS, O’Connor DH (2008) Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics 60:37–46
Kita YF, Hosomichi K, Kohara S, Itoh Y, Ogasawara K, Tsuchiya H, Torii R, Inoko H, Blancher A, Kulski K, Shiina T (2009) MHC class I A loci polymorphism and diversity in three Southeast Asian populations of cynomolgus macaques. Immunogenetics 61:635–648
Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y (2007) Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319–323
Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH (2005) Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol 175:5230–5239
Kulski JK, Anzai T, Shiina T, Inoko H (2004) Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol 21:2079–2091
Lang KC (2006) http://pin.primate.wisc.edu/factsheets/entry/long-tailed_macaque
Lawler JV, Endy TP, Hensley LE, Garrison A, Frinz EA, Lesar M, Baric RS, Kulesh DA, Norwood DA, Wasieloski LP, Ulrich MP, Slezak TR, Vitalis E, Hugging JW, Jahrling PB, Paragas J (2006) Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med 3:677–686
Loffredo JT, Bean AT, Beal DR, León EJ, May GE, Piaskowski SM, Furlott JR, Reed J, Musani SK, Rakasz EG, Friedrich TC, Wilson NA, Allison DB, Watkins DI (2008) Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol 82:1723–1738
Matano T, Kobayashi M, Igarashi H, Takeda A, Nakamura H, Kano M, Sugimoto C, Mori K, Iida A, Hirata T, Hasegawa M, Yuasa T, Miyazawa M, Takahashi Y, Yasunami M, Kimura A, O’Connor DH, Watkins DI, Nagai Y (2004) Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J Exp Med 199:1709–1718
Mee ET, Berry N, Ham C, Sauermann U, Maggiorella MT, Martinon F, Verschoor EJ, Heeney JL, Le Grand R, Titti F, Almond N, Rose NJ (2009) Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics 61:327–339
Naruse TK, Chen Z, Yanagida R, Yamashita T, Saito Y, Mori K, Akari H, Yasutomi Y, Miyazawa M, Matano T, Kimura A (2010) Diversity of MHC class I genes in Burmese-origin rhesus macaques. Immunogenetics 62:601–611
Otting N, deVos-Rouweler AJM, Heijmans CMC, de Groot NG, Doxiadis GG, Bontrop RE (2007) MHC class I A region diversity and polymorphism in macaque species. Immunogenetics 59:367–375
Otting N, Heijmans CM, van der Wiel M, de Groot NG, Doxiadis GG, Bontrop RE (2008) A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics 60:507–514
Otting N, Doxiadis GG, Bontrop RE (2009) Definition of Mafa-A and -B haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 61:745–753
Pendley CJ, Becker EA, Karl JA, Blasky AJ, Wiseman RW, Hughes AL, O’Connor SL, O’connor DH (2008) MHC class I characterization of Indonesian cynomolgus macaques. Immunogenetics 60:339–351
Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SGE (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucl Acid Res 31:311–314
Rosner C, Kruse PH, Lübke T, Walter L (2010) Rhesus macaque MHC class I molecules show differential subcellular localization. Immunogenetics 62:149–158
Slukvin II, Lunn DD, Watkins DI, Golos TG (2000) Placental expression of the nonclassical MHC class I molecule Mamu-AG at implantation in the rhesus monkey. Proc Natl Acad Sci USA 97:9104–9109
Tanaka-Takahashi Y, Yasunami M, Naruse T, Hinohara K, Matano T, Mori K, Miyazawa M, Honda M, Yasutomi Y, Nagai Y, Kimura A (2007) Reference strand-mediated conformation analysis-based typing of multiple alleles in the rhesus macaque MHC class I Mamu-A and Mamu-B loci. Electrophoresis 28:918–924
Tsukamoto T, Dohki S, Ueno T, Kawada M, Takeda A, Yasunami M, Naruse T, Kimura A, Takiguchi M, Matano T (2008) Determination of a major histocompatibility complex class I restricting simian immunodeficiency virus Gag241-249 epitope. AIDS 22:993–998
Uda A, Tanabayashi K, Yamada YK, Akari H, Lee YJ, Mukai R, Terao K, Yamada A (2004) Detection of 14 alleles derived from the MHC class I A locus in cynomolgus monkeys. Immunogenetics 56:155–163
Uda A, Tanabayashi K, Fujita O, Hotta A, Terao K, Yamada A (2005) Identification of the MHC class I B locus in cynomolgus monkeys. Immunogenetics 57:189–197
Urvater JA, Otting N, Loehrke JH, Rudersdorf R, Slukvin II, Piekarcyk MS, Golos TG, Hughes AL, Bontrop RE, Watkins DI (2000) Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J Immunol 164:1386–1398
Watanabe A, Shiina T, Shimizu S, Hosomichi K, Yanagiya K, Kita YF, Kimura T, Soeda E, Torii R, Ogasawara K, Kulski JK, Inoko H (2006) A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics 89:402–412
Wiseman RW, O’Connor DH (2007) Major histocompatibility complex-defined macaques in transplantation research. Transplant Rev 21:17–25
Wiseman RW, Wojcechowskyj JA, Greene JM, Blasly AJ, GoponT SomaT, FriedrichT C, O’Connor SL, O’Connor DH (2007) Simian immunodeficiency virus SIV mac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol 81:349–361
Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E Jr, Wright C, Harkins T, O’Connor DH (2009) Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med 15:1322–1326
Wu J, Bassinger S, Montoya GD, Chavez L, Jones CE, Holder-Lockyer B, Masten B, Williams TM, Prilliman KR (2008) Allelic diversity within the high frequency Mamu-A2*05/Mane-A2*05(Mane-A*06)/Mafa-A2*05 family of macaque MHC-A loci. Tissue Antigens 72:29–38
Acknowledgments
We thank Ms. Yukiko Ueda for her technical assistance. We acknowledge Dr. Natasja de Groot and Dr. Nel Otting for assigning nomenclature of Mafa class I alleles. This work was supported in part by research grants from the Ministry of Health, Labor and Welfare, Japan; the Japan Health Science Foundation; the program of Founding Research Centers for Emerging and Reemerging Infection Disease; the program of Research on Publicly Essential Drugs and Medical Devices; and Grant-in-Aids for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan. This work was also supported by a program of support for women researchers from the Tokyo Medical and Dental University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yusuke Saito and Taeko K. Naruse contributed equally to this work.
Rights and permissions
About this article
Cite this article
Saito, Y., Naruse, T.K., Akari, H. et al. Diversity of MHC class I haplotypes in cynomolgus macaques. Immunogenetics 64, 131–141 (2012). https://doi.org/10.1007/s00251-011-0568-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-011-0568-y