Abstract
The major histocompatibility complex (MHC) class I B gene/allelic repertoire was investigated in a pedigreed population of cynomolgus macaques of mixed Indonesian/Malaysian origin. The Mafa-B alleles detected in this cohort are mostly specific for a given geographic area, and only a small number of alleles appears to be shared with other populations. This suggests the fast evolution of Mafa-B alleles due to adaptation to new environments. In contrast to humans, the B locus in Old World monkeys displays extensive copy number variation. The Mafa-B and previously defined -A gene combinations segregate in families and thus allowed the definition of extended haplotypes. In many cases it was possible to assign a particular Mafa-I allele to one of these Mafa-A/B haplotypes as well. The presence of a large number of stable haplotypes in this cohort of animals, which was pedigreed for up to eight generations, looks promising for developing discriminative MHC typing tools that are less cumbersome. Furthermore, the discovery of 53 unreported Mafa-B sequences expands the lexicon of alleles significantly, and may help in understanding the complex organisation of the macaque B region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cynomolgus macaque (Macaca fascicularis), also known as the crab-eating or long-tailed macaque, is widely used as an animal model in biomedical studies. Currently this species is applied as often as the commonly used rhesus macaque (Macaca mulatta). Cynomolgus monkeys are used as models for infectious diseases, such as AIDS, SARS and tuberculosis, as well as for transplantation research (McAuliffe et al. 2004; Wiseman et al. 2007; Aoyama et al. 2009; Mee et al. 2009; Reed et al. 2009). Owing to use of macaques in immune-related research, thorough investigations of their major histocompatibility complexes (MHC) are required. The MHC represents a multigene family in which the proteins play a key role in the generation of adaptive immune responses in vertebrate species. The class I and II genes of the MHC display abundant polymorphism that has a profound impact on features such as disease susceptibility, organ transplantation, and reproduction success.
The MHC systems in humans (HLA) and in other primate species have been studied extensively (Bontrop 2006). The orthologues of the classical HLA-A and -B genes, which are involved in the presentation of intracellularly processed peptides to cytotoxic T cells, are present in the rhesus and cynomolgus macaque (Boyson et al. 1996; Krebs et al. 2005). However, in these animals the genes have undergone several rounds of duplication and display copy number variation (Anzai et al. 2003; Daza-Vamenta et al. 2004; Otting et al. 2005). Whereas in humans only one copy of the HLA-A and -B genes is present, in macaques seven A-like genes are distinguished. On each haplotype, one polymorphic gene is observed, named Mamu-A1 or Mafa-A1, in combination with one or two oligomorphic genes designated Mamu- or Mafa-A2 up to -A7, respectively (Otting et al. 2007; Pendley et al. 2008; Campbell et al. 2009; Kita et al. 2009). The same organisation is also applicable to the pig-tailed macaque (Macaca nemestrina) (Lafont et al. 2007; Wu et al. 2008).
The situation for the HLA-B orthologues in macaque species is even more complicated. In one rhesus macaque, the MHC region was completely sequenced, yielding one complete haplotype of 5.3 megabase-pairs. On this haplotype, 19 distinct Mamu-B genes were present, of which 14 genes have the potential to code for bonafide proteins (Anzai et al. 2003; Daza-Vamenta et al. 2004; Bonhomme et al. 2008; Doxiadis et al. 2009). For the MHC of cynomolgus macaque a BAC-based contig map was constructed (Watanabe et al. 2007). Although the degree of gene multiplication is less than in the rhesus macaque, this contig map still contains 12 distinct Mafa-B like loci. Sequencing studies at the cDNA level, however, have shown that only two or three genes per haplotype are transcribed at considerable levels (majors) in rhesus- and in cynomolgus macaques (Krebs et al. 2005; Otting et al. 2005, 2008; Pendley et al. 2008). At least one other B-like gene, characterised by low levels of polymorphism and transcription (minors), is present on all haplotypes. It has been designated Mamu-I, Mafa-I, and Mane-I in the respective species of macaques (Urvater et al. 2000; Robinson et al. 2003). On the completely sequenced MHC-region of the rhesus macaque, this locus is designated as the Mamu-B3 gene (Daza-Vamenta et al. 2004; Doxiadis et al. 2009).
The sequencing of macaques from different geographic areas has shown that each population has its own characteristic set of Mamu/Mafa-A and -B alleles, and only a few alleles are shared between cohorts/populations (Krebs et al. 2005; Otting et al. 2008; Campbell et al. 2009). This is in contrast to the data that were observed for the MHC class II sequences obtained from these species (Otting et al. 2002; Doxiadis et al. 2006; O’Connor et al. 2007; de Groot et al. 2008). Moreover, the interspecies sharing of MHC class I alleles in rhesus and cynomolgus macaques is in the same order of magnitude as the intraspecies sharing (Otting et al. 2007).
We have access to cynomolgus macaques that have been pedigreed for eight generations, and the origin of the animals was determined based on mtDNA analyses. In an earlier study, we showed that the Mafa-A alleles are mostly unique for this population. Hence, a unique set of Mafa-B alleles is expected to be present in the same animals. The question is whether these B-alleles segregate in a stable linkage to the already described Mafa-A sequences in these animals. Should this be the case, MHC typing on this cohort may be then performed using less cumbersome techniques: for instance, those based on microsatellite or SNP analyses.
Furthermore, expanding the lexicon of Mafa-B alleles may provide more insight into the organisation of the macaque B-region, and may help in the definition of different lineages and loci, resulting in a more appropriate nomenclature.
Materials and methods
Animals and cell lines
The cynomolgus macaques used in this study had originally been kept at the University of Utrecht, where the animals were housed in social groups for up to eight generations. Recently, however, the colony of 135 animals was transferred to the new facilities at the Biomedical Primate Research Centre (BPRC), for the purpose of behavioural studies. The BPRC had access to blood samples drawn during health-checks, and lymphoblastoid cell-lines were established. The origin of the animals was determined by mitochondrial DNA (12S rRNA) analyses (Doxiadis et al. 2003; de Groot et al. 2008), and the founder animals appear to have originated either in the Indonesian islands or in continental Malaysia.
cDNA, cloning, and sequencing
For all animals used in this study RNA was isolated from lymphoblastoid B-cells (Rneasy kit, Qiagen) and subjected to One-Step RT-PCR, as recommended by the supplier (Qiagen or Promega). The primers 5′MBS: AATTCATGGCGCCCCGAACCCTCCTCCTGC and 3′MBS: CTAGACCACACAAGACAGTTGTCTCAG were used that anneal specifically to Mhc-B transcripts in macaques (Boyson et al. 1996). Furthermore, for a subset of the animals the generic class I primers 5′ GGACTCAGAATCTCCCCAGACGCCGAG and 3′ TCTCAGTCCCTCACAAGGCAGCTGTC were used. The final elongation step was extended to 30 min to generate a 3′dA overhang. The RT-PCR products were cloned using the InsT/Aclone kit (Fermentas) or the PCR cloning kit (Qiagen). After transformation, 32 to 48 colonies were picked for plasmid isolation. Sequencing reactions were performed using the BigDye terminator cycle sequencing kit, and samples were run on an automated capillary sequencing system (Applied Biosystems Genetic Analyzer 3100).
Phylogenetic analyses and nomenclature
Sequences were analysed using Sequence Navigator Software version 1.0.1 (Applied Biosystems) and MacVector™ version 10.6.0 (Oxford Molecular Group), followed by manual adjustments. After the alignments of all Mamu- and Mafa-B exon 1–4 sequences using the MacVector software, version 10.6.0, phylogenetic analysis was performed with the phylogeny.fr pipeline (Dereeper et al. 2008) using maximum likelihood (ML) of the software PhyML 3.0 with the substitution model HKY85 with 4 categories, the gamma shape parameter of 0.407, a transition/transversion ratio of 2.130, and a SH-like approximate Likelihood-Ratio Test (aLRT) for statistical test of branch support. For tree rendering the pipeline uses the program TreeDyn 198, and the output tree is rooted using the mid-point rooting method.
The Mafa-B alleles have been named according to published nomenclature proposals (Klein et al. 1990; Ellis et al. 2006); however, the number of Mafa-B lineages has exceeded 100, and for the lineage numbers three digits have been introduced. In this document, the designations that were published previously are extended with a zero, and contain five ciphers. Two alleles that are highly similar receive the same lineage number, but any difference is indicated by the allele number (fourth and fifth cipher). If two alleles have synonymous base-pair differences, they receive an identical allele number, and the difference is indicated by a sixth and seventh digit. For example, Mafa-B*0950101 has synonymous differences in comparison to Mafa-B*0950102 and non-synonymous ones as compared to Mafa-B*09502. The novel alleles were submitted to the EMBL-EBI database (accession numbers FM212793-FM212843, FM246485-FM246500, FN423784, FN546179, and FN546180) and to the Non-human primate section of the IMGT/MHC Immuno Polymorphism Database (Robinson et al. 2003).
Results and discussion
Mafa-B, -I and -A sequences
In the cohort of 115 cynomolgus macaques, three to eight different Mafa-B sequences per animal were detected, with varying levels of transcription. The sequences, of which at least three identical clones were present, were reported as alleles. In most cases, these alleles were also confirmed in different animals. In total, 69 Mafa-B alleles were present, of which 16 were previously described by other research groups (Uda et al. 2005; Lafont et al. 2007; Pendley et al. 2008; Wu et al. 2008; Campbell et al. 2009; Kita et al. 2009). The other 53 sequences have been submitted to EMBL-EBI and to the MHC-NHP database (Robinson et al. 2003), and have been catalogued. A list of the Mafa-B alleles is provided, including the accession numbers, and the reference animals (Table 1).
In most animals, sequences that are alleles of the Mafa-I gene, the equivalent of the oligomorphic Mamu-I locus, were detected (Urvater et al. 2000). The Mamu-I/Mafa-I locus has the characteristics of a nonclassical, with low levels of polymorphism and transcription. Only 12 Mafa-I sequences met the criterion of three identical clones, and eight of them have been submitted as novel alleles (Table 1). Nevertheless, the Mafa-I gene appears to be present on all haplotypes.
The Mhc-A region-derived class I alleles of the animals in this cohort were sequenced in an earlier study (Otting et al. 2007). In those analyses, primers were used that are specific for Mhc-A alleles in macaques. However, in some animals the Mafa-A1 locus was not amplified by this primer set. In the present study, RT-PCR was performed with macaque Mhc-B-specific primers, and with the generic class I primers for those animals previously lacking a Mafa-A1 sequence. The use of these generic primers resulted in the detection of six new alleles for the Mafa-A1 locus, and one for both the Mafa-A2 and the -A5 genes (Table 1), and as such extends the earlier reported data.
Mafa-A, -B, and -I haplotypes
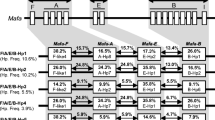
Since pedigree data were available, it was possible to determine the combinations of Mafa-B alleles on one chromosome (haplotype). Most haplotypes contain two major alleles, in combination with one or two alleles with lower levels of transcription, or minors, as based on the number of picked clones within a PCR sample. Unfortunately, it can not be excluded that some alleles are incorrectly considered as minors due to primer inconsistencies. Haplotypes with only one or three majors were also observed. Moreover, it was possible to extend these Mafa-B combinations with specific -A region configurations/haplotypes that were described in an earlier study (Otting et al. 2007). Combinations of Mafa-A and -B alleles that are segregating, and are observed in at least two related animals are listed (Table 2). Although more than one Mafa-A gene is present on the cynomolgus chromosome, only alleles of the highly polymorphic Mafa-A1 locus are provided for sake of convenience. Six additional Mafa-A/B haplotypes were seen in only one animal, whereas nine sequence combinations were ambiguous, and are not listed in the table. Further analyses on these animals, and on their offspring are needed to find out if these are recombinations.
The number of at least 24 distinct haplotypes is high, though they appear to be stable entities in this population of macaques; recombination was seldom observed. Only one case of crossing over between the Mhc-A and -B region is seen; Mafa-A1*03101 is present in conjunction with two different Mafa-B combinations (7 and 8 in Table 2). For eleven of the 24 Mafa-A/-B haplotypes it was possible to add an associated Mafa-I allele. Preliminary studies with DRB-microsatellites (Doxiadis et al. 2007; de Groot et al. 2008) indicate that the Mafa-A/B haplotypes are also linked to DRB-STR patterns. Further investigation should reveal whether in the future these animals and their offspring can be typed for the class I alleles by means of this extremely fast and accurate typing technique.
In our cohort of animals, 16 Mafa-B alleles were detected that were already described in studies on other cynomolgus populations. To determine whether these alleles were arranged in haplotypes that are shared between populations a comparison was made. Three of these combinations were observed. Pendley and coworkers have already described the sharing of B*01201/B*05001/2 and B*04501/B*05101 allele-combinations in Indonesian and Mauritian cohorts (Pendley et al. 2008), which illustrates that the Mauritian animals originate in the archipelago. Probably the animals were introduced to the island by merchant ships in the Dutch Golden Age (Sussman and Tattersall 1986). Both haplotypes are present in our animals, and are listed, respectively, as 13 and 15 in Table 2. The first one was extended to A1*06001/B*0110102/B*01201/B*05001. In Pendley’s cohort this combination of Mafa-B alleles is seen in an animal that also transcribes Mafa-A1*06003. This allele differs by two basepairs from Mafa-A1*06001. With genotyping based on reference-strand conformational analysis (RSCA), Krebs and co-workers found the combination B*0430101/B*0440101/B*0460101 in a cohort of Mauritian animals (Krebs et al. 2005). In our animals, however, we observed this set without B*0430101 (Table 2, haplotype 14). It is possible that this allele is a minor, and therefore was probably missed in our cloning procedures. In the RSCA study the B*0430101 peak is also low in comparison to the peaks of the other two alleles. The three shared haplotypes were present in cohorts that, like most of our animals at the BPRC, originate in the Indonesian Islands. Sharing of haplotypes with the recently described Filipino cynomolgus macaques was not observed (Campbell et al. 2009).
The restricted sharing of alleles in different populations of cynomolgus macaques, and moreover the recombination of similar alleles into other haplotypes in these populations, suggests that the diversity within the Mafa-B region has been the result of recombination and reshuffling of B-like loci during evolution. The cohort under study has been pedigreed for up to 8 generations, however, the haplotypes listed are observed in maximal five generations of related animals. The finding that within this cohort only one crossing between Mafa-A and Mafa-B is observed may be due to this relatively small number of generations. The fact that each cohort of animals has its own set of alleles and haplotypes necessitates investigation of the MHC for each cohort under study. Only within a breeding colony are the haplotypes more or less stable and predictable, and once the haplotypes are inventoried, robust MHC typing based on microsatellite analyses may be performed. Recombinations can be traced by using microsatellites spanning the whole MHC region.
Comparison to rhesus macaques
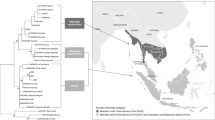
To investigate the presence of shared alleles between cynomolgus and rhesus macaques, phylogenetic analyses were performed on all 182 Mafa-B and 206 Mamu-B sequences, published thus far. Only transcribed alleles, based on analyses of cDNA were included. A subset of these analyses, comprising the exons 2, 3 and 4 of the alleles detected in the present cynomolgus macaques and alleles of known haplotypes in rhesus macaques, is displayed in Fig. 1. The phylogenetic tree shows that cynomolgus and rhesus sequences are fully intertwined, and each clade contains alleles of both species. In total, 17 sets of alleles were observed that are identical for all exons (Table 3), and this number is in the same order of magnitude as the shared alleles among different populations of cynomolgus macaques. For instance, 25 Mafa-B alleles were described in the cohort Filipino cynomolgus macaques, of which three alleles were shared with animals of Indonesian origin (Campbell et al. 2009). Next to these shared alleles are several that differ by only one or two basepairs. Identity at the predicted amino-acid level for these alleles was not investigated.
Phylogenetic analyses of Mafa-B alleles detected in this study and Mamu-B alleles in known haplotypes of Indian and Chinese rhesus monkeys. The analyses are based on the exon 2, 3 and 4 sequences. Alleles that seem identical in this three may have basepair differences in other exons. The two species of macaques are indicated by different colors. Mafa/Mamu-B alleles of the one shared haplotype are depicted in green. The extension of allele-names with mi means that these are minors; alleles with relatively low transcription
The sharing of haplotypes between cynomolgus and rhesus macaques was also investigated. Only one combination of three Mafa-B sequences was present in the rhesus macaque, and interestingly this was the B*0110102/B*01201/B*05001 combination mentioned above. Moreover, this Mamu-B*03601/2/B*04501/2/B*03701 haplotype is one of four combinations that is shared by Indian and Chinese rhesus macaques, apart from a few basepair differences. The cynomolgus version differs by three basepairs in Mafa-B*01201 from the Indian rhesus macaque haplotype. The presence of the haplotype in different cohorts of rhesus monkeys and in the Indonesian/Mauritian cynomolgus macaques suggests that its ancestor was already present before the separation of both species. The stability of the haplotype during macaque evolution may have been caused by a significant advantage in the combat of intercellular pathogens. It is also possible that the sharing of the haplotype results from hybridisation. Molecular studies have revealed that introgression from rhesus macaques to cynomolgus monkeys has occured into the Indo-Chinese peninsula (Bonhomme et al. 2009).
Similarities in the genetics of the macaque MHC class I regions are at this stage unfortunately not reflected in the nomenclature for Mhc-B alleles. The recent renaming of Mhc-A alleles has led to a nomenclature in which the distinct A loci and lineage numbers are compatible for all macaque species. For the Mhc-B region in macaques, however, it is not yet possible to assign the transcribed alleles to distinct B loci on the chromosome (e.g. Mamu-B1, -B2, -B3 etc.). However, it would be useful to adjust the lineage numbers (first three ciphers after the asterisk) so that the same numbers refer to similar sequences in different macaque species. Should more information become available in the near future on the number and order of B genes/loci on the macaque haplotypes, the allele designations may then easily be extended by a cipher following the B, without affecting the lineage numbers.
References
Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, Yamagata T, Kulski JK, Naruse TK, Fujimori Y, Fukuzumi Y, Yamazaki M, Tashiro H, Iwamoto C, Umehara Y, Imanishi T, Meyer A, Ikeo K, Gojobori T, Bahram S, Inoko H (2003) Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci U S A 100:7708–7713
Aoyama A, Ng CY, Millington TM, Boskovic S, Murakami T, Wain JC, Houser SL, Madsen JC, Kawai T, Allan JS (2009) Comparison of lung and kidney allografts in induction of tolerance by a mixed-chimerism approach in cynomolgus monkeys. Transplant Proc 41:429–430
Bonhomme M, Doxiadis GG, Heijmans CM, Vervoort V, Otting N, Bontrop RE, Crouau-Roy B (2008) Genomic plasticity of the immune-related Mhc class I B region in macaque species. BMC Genomics 9:514
Bonhomme M, Cuartero S, Blancher A, Crouau-Roy B (2009) Assessing natural introgression in 2 biomedical model species, the rhesus macaque (Macaca mulatta) and the long-tailed macaque (Macaca fascicularis). J Hered 100:158–169
Bontrop RE (2006) Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum Immunol 67:388–397
Boyson JE, Shufflebotham C, Cadavid LF, Urvater JA, Knapp LA, Hughes AL, Watkins DI (1996) The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol 156:4656–4665
Campbell KJ, Detmer AM, Karl JA, Wiseman RW, Blasky AJ, Hughes AL, Bimber BN, O’Connor SL, O’Connor DH (2009) Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics 61:177–187
Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE (2004) Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 14:1501–1515
de Groot N, Doxiadis GG, de Vos-Rouweler AJ, de Groot NG, Verschoor EJ, Bontrop RE (2008) Comparative genetics of a highly divergent DRB microsatellite in different macaque species. Immunogenetics 60:737–748
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–469
Doxiadis GG, Otting N, de Groot NG, de Groot N, Rouweler AJ, Noort R, Verschoor EJ, Bontjer I, Bontrop RE (2003) Evolutionary stability of MHC class II haplotypes in diverse rhesus macaque populations. Immunogenetics 55:540–551
Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE (2006) Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics 58:259–268
Doxiadis GG, de Groot N, Claas FH, Doxiadis II, van Rood JJ, Bontrop RE (2007) A highly divergent microsatellite facilitating fast and accurate DRB haplotyping in humans and rhesus macaques. Proc Natl Acad Sci U S A 104:8907–8912
Doxiadis GG, Heijmans CM, Bonhomme M, Otting N, Crouau-Roy B, Bontrop RE (2009) Compound evolutionary history of the rhesus macaque MHC class I B region revealed by microsatellite analysis and localization of retroviral sequences. PLoS One 4:e4287
Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, Kennedy LJ, Robinson J, Smith DM, Stear MJ, Stet RJ, Waller MJ, Walter L, Marsh SG (2006) ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics 57:953–958
Kita YF, Hosomichi K, Kohara S, Itoh Y, Ogasawara K, Tsuchiya H, Torii R, Inoko H, Blancher A, Kulski JK, Shiina T (2009) MHC class I A loci polymorphism and diversity in three Southeast Asian populations of cynomolgus macaque. Immunogenetics 61:635–648
Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217–219
Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH (2005) Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol 175:5230–5239
Lafont BA, McGraw CM, Stukes SA, Buckler-White A, Plishka RJ, Byrum RA, Hirsch VM, Martin MA (2007) The locus encoding an oligomorphic family of MHC-A alleles (Mane-A*06/Mamu-A*05) is present at high frequency in several macaque species. Immunogenetics 59:211–223
McAuliffe J, Vogel L, Roberts A, Fahle G, Fischer S, Shieh WJ, Butler E, Zaki S, St Claire M, Murphy B, Subbarao K (2004) Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 330:8–15
Mee ET, Berry N, Ham C, Sauermann U, Maggiorella MT, Martinon F, Verschoor EJ, Heeney JL, Le Grand R, Titti F, Almond N, Rose NJ (2009) Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics 61:327–339
O’Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, O’Connor DH (2007) Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 59:449–462
Otting N, de Groot NG, Doxiadis GG, Bontrop RE (2002) Extensive Mhc-DQB variation in humans and non-human primate species. Immunogenetics 54:230–239
Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE (2005) Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A 102:1626–1631
Otting N, de Vos-Rouweler AJ, Heijmans CM, de Groot NG, Doxiadis GG, Bontrop RE (2007) MHC class I A region diversity and polymorphism in macaque species. Immunogenetics 59:367–375
Otting N, Heijmans CM, van der Wiel M, de Groot NG, Doxiadis GG, Bontrop RE (2008) A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics 60:507–514
Pendley CJ, Becker EA, Karl JA, Blasky AJ, Wiseman RW, Hughes AL, O’Connor SL, O’Connor DH (2008) MHC class I characterization of Indonesian cynomolgus macaques. Immunogenetics 60:339–351
Reed SG, Coler RN, Dalemans W, Tan EV, DeLa Cruz EC, Basaraba RJ, Orme IM, Skeiky YA, Alderson MR, Cowgill KD, Prieels JP, Abalos RM, Dubois MC, Cohen J, Mettens P, Lobet Y (2009) Defined tuberculosis vaccine, Mtb72F/AS02A, evidence of protection in cynomolgus monkeys. Proc Natl Acad Sci U S A 106:2301–2306
Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 31:311–314
Sussman R, Tattersall I (1986) Distribution, abundance, and putative ecological strategy of Macaca fascicularis on the island of Mauritius, Southwestern Indian Ocean. Folia Primatol 46:28–43
Uda A, Tanabayashi K, Fujita O, Hotta A, Terao K, Yamada A (2005) Identification of the MHC class I B locus in cynomolgus monkeys. Immunogenetics 57:189–197
Urvater JA, Otting N, Loehrke JH, Rudersdorf R, Slukvin II, Piekarczyk MS, Golos TG, Hughes AL, Bontrop RE, Watkins DI (2000) Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J Immunol 164:1386–1398
Watanabe A, Shiina T, Shimizu S, Hosomichi K, Yanagiya K, Kita YF, Kimura T, Soeda E, Torii R, Ogasawara K, Kulski JK, Inoko H (2007) A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics 89:402–412
Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O’Connor SL, O’Connor DH (2007) Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol 81:349–361
Wu J, Bassinger S, Montoya GD, Chavez L, Jones CE, Holder-Lockyer B, Masten B, Williams TM, Prilliman KR (2008) Allelic diversity within the high frequency Mamu-A2*05/Mane-A2*05 (Mane-A*06)/Mafa-A2*05 family of macaque MHC-A loci. Tissue Antigens 72:29–38
Acknowledgements
The authors wish to thank Donna Devine for editing the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study was supported in part by the National Institutes of Health, project 1-R24-RR 16038-01 (Catalog of federal assistance 93.306).
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Otting, N., Doxiadis, G.G.M. & Bontrop, R.E. Definition of Mafa-A and -B haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 61, 745–753 (2009). https://doi.org/10.1007/s00251-009-0412-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-009-0412-9