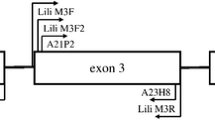
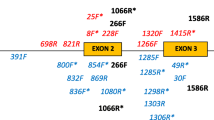
Abstract
A major challenge facing studies of major histocompatibility complex (MHC) evolution in birds is the difficulty in genotyping alleles at individual loci, and the consequent inability to investigate sequence variation and selection pressures for each gene. In this study, four MHC class I loci were isolated from the red-billed gull (Larus scopulinus), representing both the first characterized MHCI genes within Charadriiformes (shorebirds, gulls, and allies) and the first full-length MHCI sequences described outside Galloanserae (gamebirds + waterfowl). Complete multilocus genotypes were obtained for 470 individuals using a combination of reference-strand conformation analysis and direct sequencing of gene-specific amplification products, and variation of peptide-binding region (PBR) exons was surveyed for all loci. Each gene is transcribed and has conserved sequence features characteristic of antigen-presenting MHCI molecules. However, higher allelic variation, a more even allele frequency distribution, and evidence of positive selection acting on a larger number of PBR residues suggest that only one locus (Lasc-UAA) functions as a major classical MHCI gene. Lasc-UBA, with more limited variation and PBR motifs that encompass a subset of Lasc-UAA diversity, was assigned a putative minor classical function, whereas the divergent and largely invariant binding-groove motifs of Lasc-UCA and -UDA are suggestive of nonclassical loci with specialized ligand-binding roles.







Similar content being viewed by others
References
Afanassieff M, Goto RM, Ha J, Sherman MA, Zhong LW, Auffray C, Coudert F, Zoorob R, Miller MM (2001) At least one class I gene in restriction fragment pattern-Y (Rfp-Y), the second MHC gene cluster in the chicken, is transcribed, polymorphic, and shows divergent specialization in antigen binding region. J Immunol 166:3324–3333
Alcaide M, Edwards SV, Cadahía L, Negro JJ (2009) MHC class I genes of birds of prey: isolation, polymorphism and diversifying selection. Conserv Genet 10:1349–1355
Allen RL, O’Callaghan CA, McMichael AJ, Bowness P (1999) HLA-B27 can form a novel β2-microglobulin-free heavy chain homodimer structure. J Immunol 162:5045–5048
Babik W (2010) Methods for MHC genotyping in non-model vertebrates. Mol Ecol Resour 10:237–251
Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC (eds) Bioinformatics for geneticists. Wiley, Chichester, pp 289–316
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329:512–518
Dean AD, Greenwald JE (1995) Use of filtered pipet tips to elute DNA from agarose gels. Biotechniques 18:980
Edwards SV, Nusser J, Gasper J (2000) Characterization and evolution of major histocompatibility complex (MHC) genes in non-model organisms, with examples from birds. In: Baker AJ (ed) Molecular methods in ecology. Blackwell Scientific, Cambridge, pp 168–207
Ewens WJ (1972) The sampling theory of selectively neutral alleles. Theor Popul Biol 3:87–112
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Garrigan D, Hedrick PW (2003) Detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57:1707–1722
Gillingham MAF, Richardson DS, Løvlie H, Moynihan A, Worley K, Pizzari T (2009) Cryptic preference for MHC-dissimilar females in male red junglefowl, Gallus gallus. Proc R Soc Lond B 276:1083–1092
Grossberger D, Parham P (1992) Reptilian class I major histocompatibility complex genes reveal conserved elements in class I structure. Immunogenetics 36:166–174
Hála K, Chaussé AM, Bourlet Y, Lassila O, Hasler V, Auffray C (1988) Attempt to detect recombination between B-F and B-L genes within the chicken B complex by serological typing, in vitro MLR, and RFLP analyses. Immunogenetics 28:433–438
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hee CS, Gao S, Loll B, Miller MM, Uchanska-Ziegler B, Daumke O, Ziegler A (2010) Structure of a classical MHC class I molecule that binds “non-classical” ligands. PLoS Biol 8:e1000557
Hess CM, Edwards SV (2002) The evolution of the major histocompatibility complex in birds. BioScience 52:423–431
Hosomichi K, Miller MM, Goto RM, Wang Y, Suzuki S, Kulski JK, Nishibori M, Inoko H, Hanzawa K, Shiina T (2008) Contribution of mutation, recombination, and gene conversion to chicken Mhc-B haplotype diversity. J Immunol 181:3393–3399
Hughes AL, Friedman R (2008) Codon-based tests of positive selection, branch lengths, and the evolution of mammalian immune system genes. Immunogenetics 60:495–506
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170
Hughes AL, Nei M (1989) Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol Biol Evol 6:559–579
Hunt HD, Fulton JE (1998) Analysis of polymorphisms in the major expressed class I locus (B-FIV) of the chicken. Immunogenetics 47:456–467
Hunt HD, Pharr GT, Bacon LD (1994) Molecular analysis reveals MHC class I intra-locus recombination in the chicken. Immunogenetics 40:370–375
Hunt HD, Goto RM, Foster DN, Bacon LD, Miller MM (2006) At least one YMHCI molecule in the chicken is alloimmunogenic and dynamically expressed on spleen cells during development. Immunogenetics 58:297–307
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Innan H (2009) Population genetic models of duplicated genes. Genetica 137:19–37
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Kaufman J (1999) Co-evolving genes in MHC haplotypes: the “rule” for nonmammalian vertebrates? Immunogenetics 50:228–236
Kaufman J, Salomonsen J, Flajnik MF (1994) Evolutionary conservation of MHC class I and class II molecules—different yet the same. Semin Immunol 6:411–424
Kaufman J, Jacob J, Shaw I, Walker B, Milne S, Beck S, Salomonsen J (1999a) Gene organization determines evolution of function in the chicken MHC. Immunol Rev 167:101–117
Kaufman J, Milne S, Göbel TWF, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S (1999b) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401:923–925
Klein J (1986) Natural history of the major histocompatibility complex. Wiley, New York
Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217–219
Koch M, Camp S, Collen T, Avila D, Salomonsen J, Wallny H-J, van Hateren A, Hunt L, Jacob JP, Johnston F, Marston DA, Shaw I, Dunbar PR, Cerundolo V, Jones EY, Kaufman J (2007) Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity 27:885–899
Kosakovski Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW (2006) GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098
Lenz TL, Becker S (2008) Simple approach to reduce PCR artefact formation leads to reliable genotyping of MHC and other highly polymorphic loci—implications for evolutionary analysis. Gene 427:117–123
Lenz TL, Eizaguirre C, Becker S, Reusch TBH (2009) RSCA genotyping of MHC for high-throughput evolutionary studies in the model organism three-spined stickleback Gasterosteus aculeatus. BMC Evol Biol 9:57
Livant EJ, Brigati JR, Ewald SJ (2004) Diversity and locus specificity of chicken MHC B class I sequences. Anim Genet 35:18–27
Longeri M, Zanotti M, Damiani G (2002) Recombinant DRB sequences produced by mismatch repair of heteroduplexes during cloning in Escherichia coli. Eur J Immunogenet 29:517–523
McVean G, Auton A (2007) LDhat 2.1: a package for the population genetic analysis of recombination. Department of Statistics, Oxford University. http://www.stats.ox.ac.uk/∼mcvean/Ldhat/manual.pdf. Accessed 24 October 2010
Mesa CM, Thulien KJ, Moon DA, Veniamin SM, Magor KE (2004) The dominant MHC class I gene is adjacent to the polymorphic TAP2 gene in the duck, Anas platyrhynchos. Immunogenetics 56:192–203
Miller MM, Goto R, Bernot A, Zoorob R, Auffray C, Bumstead N, Briles WE (1994) Two Mhc class I and two Mhc class II genes map to the chicken Rfp-Y system outside the B complex. Proc Natl Acad Sci USA 91:4397–4401
Miller HC, Andrews-Cookson M, Daugherty CH (2007) Two patterns of variation among MHC class I loci in tuatara (Sphenodon punctatus). J Heredity 98:666–677
Mills JA (1989) Red-billed gull. In: Newton I (ed) Lifetime reproduction in birds. Academic Press Ltd., London, pp 387–404
Moon DA, Veniamin SM, Parks-Dely JA, Magor KE (2005) The MHC of the duck (Anas platyrhynchos) contains five differentially expressed class I genes. J Immunol 175:6702–6712
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Noakes MA, Reimer T, Phillips RB (2003) Genotypic characterization of an MHC class II locus in lake trout (Salvelinus namaycush) from Lake Superior by single-stranded conformational polymorphism analysis and reference strand-mediated conformational analysis. Mar Biotechnol 5:270–278
Piertney SB, Oliver MK (2006) The evolutionary ecology of the major histocompatibility complex. Heredity 96:7–21
Pratt BF, O’Connor DH, Lafont BAP, Mankowski JL, Fernandez CS, Triastuti R, Brooks AG, Kent SJ, Smith MZ (2006) MHC class I allele frequencies in pigtail macaques of diverse origin. Immunogenetics 58:995–1001
Promerová M, Albrecht T, Bryja J (2009) Extremely high MHC class I variation in a population of a long-distance migrant, the scarlet rosefinch (Carpodacus erythrinus). Immunogenetics 61:451–461
Rodgers JR, Cook RG (2005) MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol 5:459–471
Saper MA, Bjorkman PJ, Wiley DC (1991) Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. J Mol Biol 219:277–319
Sawyer SA (1999) GENECONV: a computer package for the statistical detection of gene conversion. Department of Mathematics, Washington University in St. Louis. http://www.math.wustl.edu/∼sawyer/geneconv/gconvdoc.html. Accessed 24 October 2010
Shaw I, Powell TJ, Marston DA, Baker K, van Hateren A, Riegert P, Wiles MV, Milne S, Beck S, Kaufman J (2007) Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J Immunol 178:5744–5752
Shawar SM, Vyas JM, Rodgers JR, Rich RR (1994) Antigen presentation by major histocompatibility complex class I-b molecules. Annu Rev Immunol 12:839–880
Shiina T, Shimizu S, Hosomichi K, Kohara S, Watanabe S, Hanzawa K, Beck S, Kulski JK, Inoko H (2004) Comparative genomic analysis of two avian (quail and chicken) MHC regions. J Immunol 172:6751–6763
Shiina T, Hosomichi K, Hanzawa K (2006) Comparative genomics of the poultry major histocompatibility complex. Anim Sci J 77:151–162
Shum BP, Rajalingam R, Magor KE, Azumi K, Carr WH, Dixon B, Stet RJM, Adkison MA, Hedrick RP, Parham P (1999) A divergent non-classical class I gene conserved in salmonids. Immunogenetics 49:479–490
Skjødt K, Koch C, Crone M, Simonsen M (1985) Analysis of chickens for recombination within the MHC (B-complex). Tissue Antigens 25:278–282
Stephens M, Scheet P (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet 76:449–462
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tao N, Richardson R, Bruno W, Kuiken C (2009) FindModel documentation. http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html. Accessed 24 October 2010
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
van den Elsen PJ, Gobin SJP, van Eggermond MCAJ, Peijnenburg A (1998) Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics 48:208–221
Watterson GA (1978) The homozygosity test of neutrality. Genetics 88:405–417
Westerdahl H (2007) Passerine MHC: genetic variation and disease resistance in the wild. J Ornithol 148:S469–S477
Westerdahl H, Wittzell H, von Schantz T, Bensch S (2004) MHC class I typing in a songbird with numerous loci and high polymorphism using motif-specific PCR and DGGE. Heredity 92:534–542
Wilson DJ, McVean G (2006) Estimating diversifying selection and functional constraint in the presence of recombination. Genetics 172:1411–1425
Worley K, Gillingham M, Jensen P, Kennedy LJ, Pizzari T, Kaufman J, Richardson DS (2008) Single locus typing of MHC class I and class II B loci in a population of red jungle fowl. Immunogenetics 60:233–247
Yang Z, Nielsen R, Goldman N, Pederson AMK (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Acknowledgments
We thank Kristen Choffe for her help in implementing the RSCA methodology, and three anonymous reviewers for their thoughtful comments regarding an earlier version of this manuscript. Funding was provided by a Marsden grant from the Royal Society of New Zealand (JAM and AJB), the Royal Ontario Museum Governors Fund and the Natural Sciences and Engineering Research Council of Canada (AJB), and a NSERC PGS-D scholarship (AC).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. S1
Reference-strand conformation analysis. a PCRs are used to amplify a FLR allele and the same region from a test sample. PCR products are shown as double-stranded DNA molecules, with different colours indicating different alleles. A star symbol represents fluorescence incorporated into one FLR strand. b PCR products are pooled, denatured, and cooled to allow re-annealing of complementary strands. Heteroduplex DNA molecules show bulges in regions of nucleotide sequence mismatch. c Hybridized DNA is separated by polyacrylamide gel electrophoresis under nondenaturing conditions. Molecules containing labelled reference strands are visualized, with relative mobility shifts resulting from the retardation of fragments due to mismatch bulges. (PDF 120 kb)
Supplemental Fig. S2
Exon–intron organization of red-billed gull MHCI genes compared to other avian taxa. Note that no information is available for Lasc-UDA exon and intron 1. Exons are drawn as shaded boxes, and introns as connecting lines. Sequence sources: duck (A. platyrhynchos Anpl-UAA, AY885227), goose (Anser sp. isolate G5, AY387655), chicken (G. gallus BF2, AL023516), quail (C. japonica Coja-B1, AB078884), turkey (Meleagris gallopavo class I antigen alpha chain 2, DQ993255). (PDF 162 kb)
Supplemental Fig. S3
Example of RSCA screening, showing mobility shifts caused by hybridization with FLR-UAA*03 (left panes) and FLR-UAA*04 (right panes) for the same individual. a Simultaneous screening of MHCI loci. b Lasc-UCA gene-specific RSCA. c Lasc-UDA gene-specific RSCA. d RSCA of cloned alleles, with identities indicated beneath sample plots. Grey shading indicates sizing bins used in allele assignment. Arrows indicate an allele not isolated during initial cloning experiments that requires validation by gene-specific PCR assays and segregation analyses (subsequently established as allele UAA*02). (PDF 193 kb)
Supplemental Fig. S4
Sequence logos of predicted PBR residues. X-axis residue numbering corresponds to Fig. 2, and residues predicted to contact peptide mainchain atoms are marked with asterisks. (PDF 245 kb)
Supplemental Fig. S5
Inferred interlocus gene conversion in red-billed gull MHCI genes. Nucleotide sequences are shown aligned to a majority rule consensus sequence. Predicted gene conversion tracts with simulation p values <0.05 are boxed in red (UAA–UBA), blue (between UAA, UCA, and UDA), and green (UBA–UDA). Additional homogenized tracts with p > 0.05 are boxed in grey. (PDF 151 kb)
Rights and permissions
About this article
Cite this article
Cloutier, A., Mills, J.A. & Baker, A.J. Characterization and locus-specific typing of MHC class I genes in the red-billed gull (Larus scopulinus) provides evidence for major, minor, and nonclassical loci. Immunogenetics 63, 377–394 (2011). https://doi.org/10.1007/s00251-011-0516-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-011-0516-x