Abstract
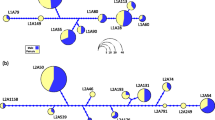
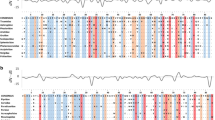
The major histocompatibility complex (MHC) is critical to host-pathogen interactions. Class II MHC is a heterodimer, with α and β subunits encoded by different genes. The peptide-binding groove is formed by the first domain of both subunits (α1 and β1), but studies of class II variation or natural selection focus primarily on the β subunit and II B genes. We explored MHC II A in Leach’s storm-petrel, a seabird with two expressed, polymorphic II B genes. We found two II A genes, Ocle-DAA and Ocle-DBA, in contrast to the single II A gene in chicken and duck. In exon 2 which encodes the α1 domain, the storm-petrel II A genes differed strongly from each other but showed little within-gene polymorphism in 30 individuals: just one Ocle-DAA allele, and three Ocle-DBA alleles differing from each other by single non-synonymous substitutions. In a comparable sample, the two II B genes had nine markedly diverged alleles each. Differences between the α1 domains of Ocle-DAA and Ocle-DBA showed signatures of positive selection, but mainly at non-peptide-binding site (PBS) positions. In contrast, positive selection within and between the II B genes corresponded to putative PBS codons. Phylogenetic analysis of the conserved α2 domain did not reveal deep or well-supported lineages of II A genes in birds, in contrast to the pronounced differentiation of DQA, DPA, and DRA isotypes in mammals. This uncertain homology complicates efforts to compare levels of functional variation and modes of evolution of II A genes across taxa.







Similar content being viewed by others
References
Aarnink A, Estrade L, Apoil PA, Kita YF, Saitou N, Shiina T, Blancher A (2010) Study of cynomolgus monkey (Macaca fascicularis) DRA polymorphism in four populations. Immunogenetics 62:123–136
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16:363–377
Bicknell AWJ, Knight ME, Bilton D, Reid JB, Burke T, Votier SC (2012) Population genetic structure and long-distance dispersal among seabird populations: implications for colony persistence. Mol Ecol 21:2863–2876
BirdLife International I (2017) Hydrobates leucorhous (amended version of 2016 assessment). The IUCN Red List of Threatened Species 2017
Bontrop RE, Otting N, De Groot NG, Doxiadis GGM (1999) Major histocompatibility complex class II polymorphisms in primates. Immunol Rev 167:339–350
Bracamonte SE, Baltazar-Soares M, Eizaguirre C (2015) Characterization of MHC class II genes in the critically endangered European eel (Anguilla anguilla). Conserv Genet Resour 7:859–870
Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC (1993) Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364:33–39
Burri R, Promerová M, Goebel J, Fumagalli L (2014) PCR-based isolation of multigene families: lessons from the avian MHC class IIB. Mol Ecol Resour 14:778–788
Burri R, Salamin N, Studer RA, Roulin A, Fumagalli L (2010) Adaptive divergence of ancient gene duplicates in the avian MHC class II β. Mol Biol Evol 27:2360–2374
Chen LC, Lan H, Sun L, Deng YL, Tang KY, Wan QH (2015) Genomic organization of the crested ibis MHC provides new insight into ancestral avian MHC structure. Sci Rep 5:7963
Číková D, De Bellocq JG, Baird SJE, Piálek J, Bryja J (2011) Genetic structure and contrasting selection pattern at two major histocompatibility complex genes in wild house mouse populations. Heredity 106:727–740
De Groot NG, Otting N, Doxiadis GGM, Balla-Jhagjhoorsingh SS, Heeney JL, Van Rood JJ, Gagneux P, Bontrop RE (2002) Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci U S A 99:11748–11753
Dearborn DC, Gager AB, McArthur AG, Gilmour ME, Mandzhukova E, Mauck RA (2016) Gene duplication and divergence produce divergent MHC genotypes without disassortative mating. Mol Ecol 25:4355–4367
Fair J, Paul E, Jones J (2010) Guidelines to the use of wild birds in research. Ornithological Council, Washington
Felsenstein J (2005) PHYLIP (phylogeny inference package). 3.6.5 edn. Distributed by the author, Department of Genome Sciences, University of Washington, Seattle
Flajnik MF (2018) A cold-blooded view of adaptive immunity. Nat Rev Immunol 18:438–453
Fridolfsson A-K, Ellegren H (1999) A simple and universal method for molecular sexing of non-ratite birds. J Avian Biol 30:116–121
Gao F, Chen C, Arab DA, Du Z, He Y, Ho SYW (2019) EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol 9:3891–3898
Germain RN, Bentley DM, Quill H (1985) Influence of allelic polymorphism on the assembly and surface expression of class II MHC (Ia) molecules. Cell 43:233–242
Goebel J, Promerová M, Bonadonna F, McCoy KD, Serbielle C, Strandh M, Yannic G, Burri R, Fumagalli L (2017) 100 million years of multigene family evolution: origin and evolution of the avian MHC class IIB. BMC Genomics 18:460
Gómez D, Conejeros P, Marshall SH, Consuegra S (2010) MHC evolution in three salmonid species: a comparison between class II alpha and beta genes. Immunogenetics 62:531–542
Hedd A, Pollet IL, Mauck RA, Burke CM, Mallory ML, McFarlane Tranquilla LA, Montevecchi WA, Robertson GJ, Ronconi RA, Shutler D, Wilhelm SI, Burgess NM (2018) Foraging areas, offshore habitat use, and colony overlap by incubating Leach’s storm-petrels Oceanodroma leucorhoa in the northwest Atlantic. PLoS One 13:e0194389
Hoover B, Alcaide M, Jennings S, Sin SYW, Edwards SV, Nevitt GA (2018) Ecology can inform genetics: disassortative mating contributes to MHC polymorphism in Leach’s storm-petrels (Oceanodroma leucorhoa). Mol Ecol 27:3371–3385
Hughes AL, Nei M (1989) Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci U S A 86:958–962
Hughes AL, Nei M (1990) Evolutionary relationships of class II major-histocompatibility-complex genes in mammals. Mol Biol Evol 7:491–514
Jacob JP, Milne S, Beck S, Kaufman J (2000) The major and a minor class II β chain (B-LB) gene flank the Tapasin gene in the B-F/B-L region of the chicken major histocompatibility complex. Immunogenetics 51:138–147
Jaratlerdsiri W, Isberg SR, Higgins DP, Miles LG, Gongora J (2014) Selection and trans-species polymorphism of major histocompatibility complex class II genes in the order Crocodylia. PLoS One 9:e87534
Karaiskou N, Moran P, Georgitsakis G, Abatzopoulos TJ, Triantafyllidis A (2010) High allelic variation of MHC class II alpha antigen and the role of selection in wild and cultured Sparus aurata populations. Hydrobiologia 638:11–20
Kaufman J (1999) Co-evolving genes in MHC haplotypes: the ‘rule’ for nonmammalian vertebrates? Immunogenetics 50:228–236
Kaufman J (2018) Generalists and specialists: a new view of how MHC class I molecules fight infectious pathogens. Trends Immunol 39:367–379
Klein J (1986) Natural history of the major histocompatability complex. Wiley, New York
Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217–219
Kosch TA, Bataille A, Didinger C, Eimes JA, Rodríguez-Brenes S, Ryan MJ, Waldman B (2016) Major histocompatibility complex selection dynamics in pathogen-infected túngara frog (Physalaemus pustulosus) populations. Biol Lett 12
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu Y, Kasahara M, Rumfelt LL, Flajnik MF (2002) Xenopus class II a genes: studies of genetics, polymorphism, and expression. Dev Comp Immunol 26:735–750
Liu Z, Hu DD, Shao SJ, Huang JQ, Wang JF, Yang J (2013) Polymorphisms in major histocompatibility complex class IIα genes are associated with resistance to infectious hematopoietic necrosis in rainbow trout, Oncorhynchus mykiss (Walbaum, 1792). J Appl Ichthyol 29:1234–1240
Lotteau V, Teyton L, Burroughs D, Charron D (1987) A novel HLA class II molecule (DRα-sDQβ) created by mismatched isotype pairing. Nature 329:339–341
Mauck RA, Dearborn DC, Huntington CE (2018) Annual global mean temperature explains reproductive success in a marine vertebrate from 1955 to 2010. Glob Chang Biol 24:1599–1613
May S, Zeisset I, Beebee TJC (2011) Larval fitness and immunogenetic diversity in chytrid-infected and uninfected natterjack toad (Bufo calamita) populations. Conserv Genet 12:805–811
Minias P, Pikus E, Whittingham LA, Dunn PO (2018) A global analysis of selection at the avian MHC. Evolution 72:1278–1293
Minias P, Pikus E, Whittingham LA, Dunn PO (2019) Evolution of copy number at the MHC varies across the avian tree of life. Genome Biol Evol 11:17–28
Parker A, Kaufman J (2017) What chickens might tell us about the MHC class II system. Curr Opin Immunol 46:23–29
Raymond M, Rousset F (1995) Genepop (version-1.2)—population-genetics software for exact tests and ecumenicism. J Hered 86:248–249
Ren L, Yang Z, Wang T, Sun Y, Guo Y, Zhang Z, Fei J, Bao Y, Qin T, Wang J, Huang Y, Hu X, Zhao Y, Li N (2011) Characterization of the MHC class II α-chain gene in ducks. Immunogenetics 63:667–678
Ruberti G, Sellins KS, Hill CM, Germain RN, Fathman CG, Livingstone A (1992) Presentation of antigen by mixed isotype class II molecules in normal H-2d mice. J Exp Med 175:157–162
Salomonsen J, Marston D, Avila D, Bumstead N, Johansson B, Juul-Madsen H, Olesen GD, Riegert P, Skjødt K, Vainio O, Wiles MV, Kaufman J (2003) The properties of the single chicken MHC classical class II α chain (B-LA) gene indicate an ancient origin for the DR/E-like isotype of class II molecules. Immunogenetics 55:605–614
Salzburger W, Ewing GB, Von Haeseler A (2011) The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol Ecol 20:1952–1963
Sin YW, Dugdale HL, Newman C, MacDonald DW, Burke T (2012) MHC class II genes in the European badger (Meles meles): characterization, patterns of variation, and transcription analysis. Immunogenetics 64:313–327
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc R Soc B Biol Sci 277:979–988
Stet RJ, De Vries B, Mudde K, Hermsen T, Van Heerwaarden J, Shum BP, Grimholt U (2002) Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics 54:320–331
Sutton JT, Nakagawa S, Robertson BC, Jamieson IG (2011) Disentangling the roles of natural selection and genetic drift in shaping variation at MHC immunity genes. Mol Ecol 20:4408–4420
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Taniguchi Y, Matsumoto K, Matsuda H, Yamada T, Sugiyama T, Homma K, Kaneko Y, Yamagishi S, Iwaisaki H (2014) Structure and polymorphism of the major histocompatibility complex class II region in the Japanese crested Ibis, Nipponia nippon. PLoS One 9:e108506
Traherne JA (2008) Human MHC architecture and evolution: implications for disease association studies. Int J Immunogen 35:179–192
Tsuji H, Taniguchi Y, Ishizuka S, Matsuda H, Yamada T, Naito K, Iwaisaki H (2017) Structure and polymorphisms of the major histocompatibility complex in the Oriental stork, Ciconia boyciana. Sci Rep 7:42864
Ujvari B, Belov K (2011) Major histocompatibility complex (MHC) markers in conservation biology. Int J Mol Sci 12:5168–5186
Wilson AB, Whittington CM, Bahr A (2014) High intralocus variability and interlocus recombination promote immunological diversity in a minimal major histocompatibility system. BMC Evol Biol 14:273
Xu T, Liu J, Sun Y, Zhu Z, Liu T (2016) Characterization of 40 full-length MHC class IIA functional alleles in miiuy croaker: polymorphism and positive selection. Dev Comp Immunol 55:138–143
Xu TJ, Chen SL, Zhang YX (2010) MHC class IIα gene polymorphism and its association with resistance/susceptibility to Vibrio anguillarum in Japanese flounder (Paralichthys olivaceus). Dev Comp Immunol 34:1042–1050
Yang M, Wei J, Li P, Wei S, Huang Y, Qin Q (2016) MHC class IIα polymorphisms and their association with resistance/susceptibility to Singapore grouper iridovirus (SGIV) in orange-spotted grouper, Epinephelus coioides. Aquaculture 462:10–16
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Wong WSW, Nielsen R (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Acknowledgments
We thank Bob Mauck, Frank Hailer, Erin Voss, Sabine Berzins, and Hillary Chavez for help with this project. Funding and equipment were provided by the Arthur Vining Davis Foundations and Institutional Development Awards (P20GM0103423) from the National Institute of General Medical Sciences of the National Institutes of Health. This is contribution number 277 from the Bowdoin Scientific Station on Kent Island.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
About this article
Cite this article
Rand, L.M., Woodward, C., May, R. et al. Divergence between genes but limited allelic polymorphism in two MHC class II A genes in Leach’s storm-petrels Oceanodroma leucorhoa. Immunogenetics 71, 561–573 (2019). https://doi.org/10.1007/s00251-019-01130-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-019-01130-z