Abstract
The brown planthopper (BPH), Nilaparvata lugens (Stål), is one of the most serious and destructive pests of rice, and can be found throughout the rice-growing areas of Asia. To date, more than 24 major BPH-resistance genes have been reported in several Oryza sativa ssp. indica cultivars and wild relatives. Here, we report the genetic basis of the high level of BPH resistance derived from an Indian rice cultivar, ADR52, which was previously identified as resistant to the whitebacked planthopper (Sogatella furcifera [Horváth]). An F2 population derived from a cross between ADR52 and a susceptible cultivar, Taichung 65 (T65), was used for quantitative trait locus (QTL) analysis. Antibiosis testing showed that multiple loci controlled the high level of BPH resistance in this F2 population. Further linkage analysis using backcross populations resulted in the identification of BPH-resistance (antibiosis) gene loci from ADR52. BPH25 co-segregated with marker S00310 on the distal end of the short arm of chromosome 6, and BPH26 co-segregated with marker RM5479 on the long arm of chromosome 12. To characterize the virulence of the most recently migrated BPH strain in Japan, preliminary near-isogenic lines (pre-NILs) and a preliminary pyramided line (pre-PYL) carrying BPH25 and BPH26 were evaluated. Although both pre-NILs were susceptible to the virulent BPH strain, the pre-PYL exhibited a high level of resistance. The pyramiding of resistance genes is therefore likely to be effective for increasing the durability of resistance against the new virulent BPH strain in Japan.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Insect pests are a major biotic constraint on rice production. The brown planthopper (BPH), Nilaparvata lugens (Stål) (Homoptera: Delphacidae), is one of the most serious and destructive pests of rice (Oryza sativa L.), and can be found throughout the rice-growing areas of Asia, where it causes significant yield losses every year in susceptible cultivars (Khush 1979; Sogawa et al. 2003). Heavy infestations cause complete drying and plant death, a condition known as “hopper burn”.
The severe damage and frequent outbreaks of BPH, along with the hazardous effects of pesticides, have prompted researchers to seek BPH-resistant germplasm from various sources and to utilize the resistance genes for rice improvement. Simple sequence repeat (SSR) marker loci are widely distributed in the rice genome and can be easily analyzed using the polymerase chain reaction (PCR; McCouch et al. 2002). Thus, SSR markers have been extensively used to map rice loci for disease and BPH resistance (Chen et al. 2006; Gu et al. 2004; Jairin et al. 2007a; Jena et al. 2006; Sun et al. 2005; Yang et al. 2002). To date, more than 24 major BPH-resistance genes have been reported in indica cultivars and five wild Oryza species (O. australiensis, O. eichingeri, O. latifolia, O. officinalis, and O. minuta). Several of the BPH-resistance genes have been assigned to rice chromosomes 2, 3, 4, 6, and 12 (Jena and Kim 2010).
However, the BPH resistance controlled by these major genes is not durable. Resistance conferred by Bph1 and bph2 has already been overcome by BPH biotypes 2 and 3, respectively (Pathak and Heinrichs 1982; Tanaka and Matsumura 2000). Four BPH biotypes have been identified by the International Rice Research Institute (IRRI). Biotype 1, a BPH population collected from various places in the Philippines and maintained at IRRI, is a general biotype that can survive on susceptible rice cultivars; biotype 2 can also survive on plants that carry Bph1, such as the cultivars Mudgo and IR26; and biotype 3 can also survive on plants carrying bph2, such as the cultivars ASD7, Ptb21, and IR32 (IRRI 1975). Biotype 4, from South India, can survive on plants carrying Bph1 or bph2 and has a different reaction pattern on resistant cultivars from those of the BPH biotypes maintained at IRRI (Velusamy et al. 1984). Biotypes 1, 2, and 3 are widely distributed in southeastern and eastern Asia, whereas biotype 4 occurs only in the Indian subcontinent (Khush and Brar 1991). Changes in insect biotypes and disease races are a continued threat to increased rice production. There is thus an urgent need to identify and introduce new genes for BPH resistance from diverse sources into rice.
We previously reported that highly resistant cultivars carried multiple genes for BPH resistance (Sonoda et al. 2003). The objectives of the present study were to (1) determine the genetic basis of the resistance derived from an Indian cultivar, ADR52 by quantitative trait locus (QTL) analysis (2) perform more detailed mapping of two candidate loci for BPH resistance, and (3) characterize the resistance to the most recently migrated BPH strain, which arrived in Japan in 2006 using preliminary near-isogenic lines (pre-NILs) and a preliminary pyramided line (pre-PYL) carrying these loci.
Materials and methods
Plant materials
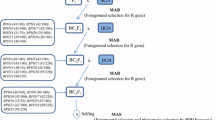
To perform the QTL analysis for resistance to BPH, we used an F2 population derived from a cross between a susceptible japonica cultivar, Taichung 65 (T65), and a resistant indica cultivar, ADR52, that was previously reported to be resistant to the whitebacked planthopper (Sogatella furcifera [Horváth]). Some F1 plants were also backcrossed with T65 to generate a BC1F1 population. A total of 10 BC1F1 plants were backcrossed with T65 as the recurrent parent to generate 10 BC2F1 populations. A total of 93 BC2F1 plants were generated, and 27 of these plants that appeared promising on the basis of marker-assisted selection (MAS) using 10 SSR markers (Table 1) were backcrossed with T65 to develop BC3F1 populations that were heterozygous for the targeted QTL regions on chromosomes 6 and 12. A total of 96 BC3F1 individuals were generated, and 12 of these that potentially carried segments of interest on chromosomes 6 and 12 were selected by MAS. The 12 selected BC3F1 plants that contained either qBPH6 or qBPH12 were self-pollinated to generate BC3F2 populations. We then analyzed two BC3F2 populations that were heterozygous for either qBPH6 (BC3F2 1) or qBPH12 (BC3F2 2) to map the BPH-resistance loci. Newly identified BPH-resistance loci were designated as BPH25 and BPH26, according to the new nomenclature system (McCouch and CGSNL 2008). These genes have been registered on-line at http://shigen.lab.nig.ac.jp/rice/oryzabase_submission/gene_nomenclature/.
We backcrossed the selected BC3F1 individuals with T65 to develop BC4F1 plants carrying the BPH-resistance genes BPH25 and BPH26 through MAS. Nine BC4F1 plants carrying the BPH-resistance genes (4 with BPH25 and BPH26, 3 with BPH25 only, and 2 with BPH26 only) were selected and self-pollinated twice to produce the BC4F3 generation. Finally, we selected plants carrying the ADR52 alleles at BPH25, BPH26, or both loci as pre-NILs and a pre-PYL from the BC4F3 population using MAS. Myint et al. (2009a) reported the lines (BC4F2) for both BPH-resistance genes and the developed lines with BPH resistance were designated as NILs or as a PYL. However, these BC4 progeny lines still have about 3% chromosomal segments from ADR52 and show delayed heading and hybrid sterility. Yara et al. (2010) demonstrated that BPH25 and BPH26 are linked to genes related to heading date and hybrid sterility, respectively. Thus, we renamed the NILs (Myint et al. 2009a) as preliminary NILs (pre-NILs) for BPH-resistance genes because of linkage drag due to the effects on heading date and hybrid sterility. In this paper, we also have changed the names of the genes and lines from bph20(t)–NIL to BPH25–pre-NIL, from Bph21(t)–NIL to BPH26–pre-NIL, and from bph20(t)/Bph21(t)–PYL to BPH25/BPH26–pre-PYL to avoid the confusion.
Insect strains
The Chikugo-89 BPH population was collected in Chikugo (Fukuoka Pref., Japan) in 1989, and the Japan-KG-06 BPH population was collected in Minamisatsuma (Kagoshima Pref., Japan) in 2006. These populations have been maintained as laboratory strains by means of continuous rearing at 25 ± 1°C under a 16-h light/8-h dark photoperiod. Long-term mass rearing of BPH in the laboratory has not affected the virulence status of these strains against cultivars with differential resistance to BPH (Myint et al. 2009b). We used the Chikugo-89 strain to map the QTLs involved in the resistance of ADR52. The two BPH strains have different virulence characteristics on cultivars with differential resistance genes (Myint et al. 2009b). Chikugo-89 is virulent against Mudgo, which carries Bph1, but avirulent against ASD7, which carries bph2. This strain, which can survive on plants that carry Bph1, shows virulence characteristics equivalent to those of biotype 2. The Japan-KG-06 strain is virulent against both Mudgo and ASD7. This strain, which can survive on plants carrying Bph1 or bph2, shows virulence characteristics equivalent to those of both biotype 2 and biotype 3. Thus, we used the Japan-KG-06 strain to monitor the current virulence status against the resistance genes BPH25 and BPH26.
Evaluation of antibiosis against BPH
For our QTL analysis, we infested 81 F2 plants at the tillering stage with ten 2nd-instar BPH nymphs. Chikugo-89 strain of BPH was used for evaluating these plants. In this test, five leaf sheaths of plants were cut off and placed in test tubes containing 10 nymphs with five replications. Nymph mortality was scored 5 days after infestation (DAI). We performed the antibiosis test in the mapping population (BC3F2) and the virulence test against the pre-NILs and the pre-PYL using the method described by Tanaka (2000). In this approach, five brachypterous females, collected within 24 h after emergence, are released onto a single plant 1 month after sowing. Antibiosis is then scored from 3 to 5 DAI. Plants with adult BPH mortality of <40% are categorized as susceptible, whereas those with adult mortality of >70% are categorized as resistant. We also examined the shape of the female’s abdomen as well as the adult mortality on the pre-NILs and the pre-PYL. Plants with females that had a heavily swollen abdomen (i.e., that produced viable eggs) or that survived for 5 days were defined as susceptible, whereas plants with females that died within 5 days or that did not have a swollen abdomen were considered to be resistant (Myint et al. 2009a).
Genotyping using SSR markers
We extracted the total DNA from fresh leaves of the parental lines, F2, BC3F2, and BC4F3 plants using the potassium acetate method (Dellaporta et al. 1983). We then determined the genotypes of the SSR loci in these plants. PCR amplification was performed in a Model 9700 PCR system (Perkin Elmer, Waltham, MA, USA). The 15-μL PCR reaction mixture contained 50 mM KCl, 10 mM Tris–HCl (pH 9.0), 1.5 mM MgCl2, 200 μM dNTPs, 0.2 μM primers, 1 U Taq polymerase (Takara, Otsu, Shiga, Japan), and 5–10 μg/mL of genomic DNA as a template. The thermal cycler was programmed for a first denaturation step of 5 min at 94°C, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with no final extension. The SSR products were resolved in 4% agarose gels by means of electrophoresis at 250 V for 1 h in 0.5× Tris–borate–EDTA buffer. The gels were stained with ethidium bromide and photographed under ultraviolet light.
Construction of a linkage map and QTL analysis
We constructed the genetic map of the F2 population using a total of 161 SSR markers distributed throughout the rice chromosomes (McCouch et al. 2002; Temnykh et al. 2001). We determined linkage of loci and map distances using Mapmaker/Exp 3.0 (Lander et al. 1987). We used the nymph mortalities and the genotypic data of the F2 population derived from the cross between T65 and ADR52 for our QTL analysis. We conducted composite interval mapping using Windows QTL Cartographer V2.0 (Wang et al. 2004). We calculated the critical threshold value of the logarithm of odds (LOD) score for QTL detection by conducting 1,000 permutation tests; this value equaled 2.9 at P < 0.05.
Mapping of genes conferring BPH resistance
We used the two BC3F2 populations for linkage analysis of the BPH-resistance genes and SSR markers. We used six SSR markers on chromosome 6 (S00310, RM6273, RM6775, MSSR1, RM8101, and RM204) and four markers (RM309, RM5479, S20103, and MSSR2) on chromosome 12 (Table 1).
Antibiosis of the most recently migrated BPH in the pre-NILs and the pre-PYL
We developed a BPH25–pre-NIL, a BPH26–pre-NIL, and a BPH25/BPH26–pre-PYL through continuous backcrossing with T65 as the recurrent parent, assisted by MAS. These lines were evaluated for BPH resistance at the tillering stage, with T65 used as a susceptible control. We evaluated the resistance to the most recently migrated BPH strain (Japan-KG-06) on the basis of the adult mortality rate and the presence or absence of a swollen abdomen 5 DAI. The data for Chikugo-89 was obtained from Myint et al. (2009a) to allow comparison of the resistance levels between the Chikugo-89 and Japan-KG-06 strains. We used seven replicates in this experiment. We analyzed the data using one-way ANOVA and performed pairwise comparisons of treatment means using the Tukey–Kramer test (SAS Institute Inc. 2003). We arcsine-transformed the adult mortality (%) before analysis.
Results
Detection of QTLs for antibiosis against BPH
We performed QTL analysis for BPH nymph mortality in the F2 population to better understand the genetic basis of BPH resistance in ADR52. The resistant parent, ADR52, produced high nymph mortality (92%). The susceptible parent, T65, showed 0% nymph mortality. The F1 plants were only moderately resistant with 61% nymph mortality. The resistance was unstable and the range of nymph mortality among replications was wide (18–92%). The F2 population exhibited a continuous frequency distribution for nymph mortality, and did not show any discrete segregation (Fig. 1). We detected three QTLs (qBPH5, qBPH6, and qBPH12), with LOD scores of 3.2, 3.5, and 3.8, on chromosomes 5, 6, and 12, respectively (Table 2, Fig. 2). We identified qBPH5 within an interval flanked by RM459 and RM3351 on the long arm of chromosome 5; this QTL contributed 17.9% of the phenotypic variation. We detected qBPH6 within an interval flanked by S00310 and RM8101 on the short arm of chromosome 6; this QTL contributed 19.1% of the phenotypic variation. We detected qBPH12 within an interval flanked by RM3331 and S20103 on the long arm of chromosome 12; this QTL contributed 15.7% of the phenotypic variation. At the qBPH6 and qBPH12 loci, the presence of alleles from the resistant parent (ADR52) significantly increased antibiosis against BPH. The allele from the susceptible parent (T65) for the remaining QTL, qBPH5, also increased antibiosis against BPH. We did not investigate qBPH5 any further in this study because it was already present in the T65 genetic background that was the recurrent backcross parent and did not increase resistance to the any BPH strains in the absence of the two new QTLs.
Location of detected QTLs, those confer resistance to the brown planthopper. Three QTLs, qBPH5, qBPH6, and qBPH12, were located on the linkage maps of rice chromosomes 5, 6, and 12, respectively. The genome-wide linkage map was constructed by 161 SSR markers using an F2 population derived from T65 × ADR52. Critical threshold value of LOD score indicated by horizontal lines was equivalent to LOD = 2.9 at an experiment-wise significance level of 0.05
Mapping of BPH-resistance genes
We developed two nearly isogenic populations (carrying qBPH6 or qBPH12) through advanced backcrossing and MAS. The two BC3F2 populations showed a clear bimodal distribution for adult mortality, which we classified into types with high and low adult mortality (Fig. 3). In population 1 (qBPH6), 16 plants showed high adult BPH mortality (100%) and 32 showed low mortality (<40%), in agreement with a 1:3 segregation ratio (χ 2 = 1.7, ns; Table 3), indicating that a single recessive gene controlled BPH resistance in this population. In population 2 (qBPH12), 41 plants showed high adult BPH mortality (>80%) and 7 showed low mortality (0%), in agreement with a 3:1 segregation ratio (χ 2 = 2.7, ns; Table 3), indicating that a single dominant gene controlled BPH resistance in this population.
These results show that the presence of a recessive gene and a dominant gene in ADR52 contributes to the high level of resistance exhibited by this cultivar by causing high adult BPH mortality. We subsequently analyzed the BC3F2 populations for BPH-resistance genes using SSR markers near the qBPH6 and qBPH12 loci. The recessive qBPH6 gene was linked with SSR marker S00310 on the distal end of the short arm of chromosome 6. Sixteen plants homozygous for the ADR52 allele at S00310 exhibited BPH resistance, with high adult mortality, whereas 32 plants that had either homozygous or heterozygous T65 alleles at S00310 were susceptible to BPH, with low adult mortality (Fig. 3a). To detect the location of the recessive gene, we conducted linkage analysis using the markers S00310, RM6273, RM6775, MSSR1, RM8101, and RM204. The genetic distances between RM8101 and the recessive gene and between RM8101 and RM204 were 1.8 and 3.2 cM, respectively (Fig. 4a). We designated the recessive gene as BPH25, according to the new nomenclature system proposed by McCouch and CGSNL (2008).
Linkage maps indicating the positions of (a) BPH25 and (b) BPH26, brown planthopper (BPH)-resistance genes on rice chromosomes 6 and 12, respectively. The framework maps at the left of the figures are quoted from Harushima et al. (1998)
The dominant qBPH12 gene co-segregated with SSR marker locus RM5479, on the long arm of chromosome 12. Forty-one plants with either homozygous or heterozygous ADR52 alleles at RM5479 exhibited BPH resistance, with high adult mortality, whereas seven plants with homozygous T65 alleles at RM5479 were susceptible to BPH, with low adult BPH mortality (Fig. 3b). To detect the location of the dominant gene, we conducted linkage analysis using the markers RM309, RM5479, S20103, and MSSR2. The genetic distances between the dominant gene and the two flanking SSR markers, RM309 and MSSR2, were 13.3 and 5.1 cM, respectively (Fig. 4b). We designated the dominant gene as BPH26.
On the basis of the markers flanking the resistance genes, we selected the pre-NILs and the pre-PYL from BC4 progeny. Four BPH25–pre-NILs that had homozygous ADR52 alleles at S00310, RM6273, RM6775, MSSR1, RM8101, and RM204 were selected. Similarly, seven BPH26–pre-NILs that had homozygous ADR52 alleles at RM309, RM5479, S20103, and MSSR2 were selected. Finally, four BPH25/BPH26–pre-PYLs that carried homozygous ADR52 alleles at six SSR markers near BPH25 and at four SSR markers near BPH26 were selected.
Characterization of resistance to the most recently migrated BPH strain (Japan-KG-06) in the pre-NILs and the pre-PYL
The resistance of the pre-NILs and the pre-PYL was evaluated using the most recently migrated BPH strain (Japan-KG-06). Very low proportions of Japan-KG-06 females died on the pre-NILs that carried a single resistance gene (14.3% for BPH25 and 11.4% for BPH26; Fig. 5a), and these proportions did not differ significantly from the proportion (8.6%) on T65, which was used to confirm susceptibility. In contrast, a high proportion of females died on the pre-PYL that carried both BPH25 and BPH26, at a level not significantly different from that on ADR52 (82.9 and 94.3%, respectively; Fig. 5a). The Japan-KG-06 females on the pre-NILs that carried only a single resistance gene developed a swollen abdomen within 5 DAI (63% for BPH25 and 86% for BPH26; Fig. 5b), at levels similar to that on T65 (86%), although the proportion was significantly smaller on the line carrying BPH25. In contrast, females on the pre-PYL that carried both genes and on ADR52 did not show a swollen abdomen (Fig. 5b).
a Adult mortality (%) of brown planthopper and b the proportion of BPH females with a swollen abdomen (%) 5 DAI on the pre-NILs and the pre-PYL of rice carrying BPH-resistance genes. The scores for Chikugo-89 strain were obtained from Myint et al. (2009a). The scores for Japan-KG-06 strain were obtained in the present study. Values represent mean and standard deviations (n = 7). Mean labeled with the same letters do not differ significantly in a and b, respectively (P < 0.01, Tukey–Kramer test)
Discussion
Our main findings are (1) the identification of two loci for BPH resistance that were derived from a highly resistant cultivar (ADR52) and (2) the effectiveness of the pre-PYL for resistance against the most recently migrated BPH strain (Japan-KG-06) in Japan. We have renamed the BPH-resistance loci according to the new gene nomenclature system for rice proposed by McCouch and CGSNL (2008) to avoid confusion among the different loci for BPH resistance. In our previous study, the genes BPH25 and BPH26 from ADR52 had been tentatively named bph20(t) and Bph21(t) (Myint et al. 2009a). However, Rahman et al. (2009) reported the same gene nomenclature in a different donor, IR71033-121-15. In addition, Bph22(t) and Bph23(t) (Ram et al. 2010) and Bph24(t) (Deen et al. 2010) have already been designated. We have therefore revised the gene names by renaming bph20(t) as BPH25 and Bph21(t) as BPH26 and have registered these gene names in Oryzabase (Yamazaki et al. 2010: http://www.shigen.nig.ac.jp/rice/oryzabase/).
A number of genes conferring resistance to BPH have been reported on the same chromosomes as BPH25 and BPH26. Among them, Bph3 and bph4 were mapped on chromosome 6. Bph3, a major resistance gene derived from the Sri Lankan cultivar Rathu Heenati, was recently mapped between markers RM19291 and RM8072 (a 190-kbp distance) on the short arm of chromosome 6 (Jairin et al. 2007b), bph4, which was derived from Babawee, was identified in nearly the same region as Bph3 (Jairin et al. 2010; Kawaguchi et al. 2001). In the present study, we mapped a recessive gene conferring resistance to BPH at SSR markers S00310, RM589, and RM588. In the Nipponbare genome sequence, RM8072 is located near the RM589 locus (Fig. 6a, b). The chromosomal location of BPH25 is very close to that of Bph3 and bph4. We tentatively named the gene BPH25 to distinguish it from the previously mapped Bph3 and bph4 genes because of the difficulty of distinguishing among the Bph3, bph4, and BPH25 loci. Fine-scale mapping of these resistance loci will offer the most promising approach to clarifying their allelic relationship.
Comparison between the chromosomal locations of (a) BPH25 (present study) and (b) Bph3 (Jairin et al. 2007b) and bph4 (Jairin et al. 2010) on the short arm of chromosome 6; and the chromosomal locations of (c) BPH26 (present study) and (d) Bph18(t) (Jena et al. 2006) and Bph1 (Cha et al. 2008) on the long arm of chromosome 12. The black arrows indicate the locations of the genes. The crosshatched bar indicates the location of the peak LOD in the QTL analysis
Rice chromosome 12 is the most important for BPH resistance, as several of its loci have been reported to confer BPH resistance: Bph1 (Hirabayashi and Ogawa 1995; Kim and Sohn 2005; Park et al. 2008; Sharma et al. 2002), bph2 (Murai et al. 2001; Murata et al. 1998; Sun et al. 2006), Bph9 (Murata et al. 2000; Su et al. 2006), Bph10(t) (Ishii et al. 1994), Bph16(t) (Hirabayashi et al. 2004), Bph18(t) (Jena et al. 2006), and Bph21(t) (Rahman et al. 2009). Comparison of the chromosomal locations between the dominant gene that we report here and the previously identified genes on the long arm of chromosome 12 was difficult because of the low-precision mapping of the loci in the previous studies, except for Bph18(t) and Bph1. The major resistance gene Bph18(t) has been identified in an introgression line (IR65482-7-216-1-2) that inherited the gene from the wild species O. australiensis. Bph18(t) was localized within a 0.843-Mb physical interval that included three BAC clones between the flanking markers RM6869 and R10289S on the long arm of chromosome 12. On the basis of gene annotation information from TIGR (http://rice.plantbiology.msu.edu/index.shtml), a sequence-tagged-site (STS) marker (7312.T4A) derived from a putative resistance gene within the BAC clone OSJNBa0028L05 was linked to Bph18(t) (Jena et al. 2006). Bph1, which was derived from Mudgo, has been identified between the STS markers pBPH4 and pBPH14 (Cha et al. 2008). These Bph1-flanking markers were located between RM6869 and R10289S, and more specifically between 7312.T4A and R10289S. Here, we mapped the dominant gene BPH26 between the SSR markers RM309 and MSSR2 at distances of 13.3 and 5.1 cM, respectively, from the two markers on the long arm of rice chromosome 12. In the Nipponbare genome sequence, RM6869 and R10289S are located between RM309 and RM5479. The chromosomal location of BPH26 is close to those of Bph18(t) and Bph1 (Fig. 6c, d). We tentatively named this gene BPH26 because of the difficulty of distinguishing among the Bph18(t), Bph1, and BPH26 loci. Map-based cloning represents the most promising approach to isolating BPH-resistance genes, and we are currently constructing a high-resolution linkage map to isolate BPH26.
Biotype selection, in which the insect overcomes host resistance as a result of selection against virulent insect genotypes, has been one of the most serious problems encountered in plant breeding for varietal resistance. The virulence of BPH populations has changed drastically since the 1970s, when commercial rice cultivars carrying the resistance genes Bph1 and bph2 were released and widely cultivated (Sogawa 1982). The BPH that immigrated into Japan became virulent against plants carrying Bph1 in the latter half of the 1980s (Sogawa 1992), and became highly virulent against rice cultivars carrying both Bph1 and bph2 in the latter half of the 1990s (Tanaka and Matsumura 2000). Myint et al. (2009b) recently confirmed the above-mentioned evidence using laboratory strains of BPH: the BPH strains collected in 1966, 1989, 1999, and 2005 in Japan showed different virulence characteristics against rice differential cultivars. They demonstrated that the 1966 BPH strain (Japan-Hatano-66) was avirulent against both Bph1 and bph2. On the other hand, the 1989 BPH strain (Japan-Chikugo-89) was virulent against Bph1 but avirulent against bph2. In addition, the 1999 BPH strain (Japan-Isahaya-99) and the 2005 BPH strain (Japan-Nishigoshi-05) were virulent against both Bph1 and bph2. Myint et al. (2009a) demonstrated that the Hatano-66 and Chikugo-89 strains were avirulent against BPH25 and BPH26 both separately and together, and that the Isahaya-99 strain was virulent against BPH25 and BPH26 separately, but avirulent against both together. The virulence pattern of the most recently migrated BPH strain in Japan (Japan-KG-06) was equivalent to that of Isahaya-99 on the basis of the adult survivorship and development of the female abdomen on the pre-NILs and the pre-PYL (Fig. 5).
Pyramiding host–plant resistance genes for insect pests is one of the most effective approaches in plant breeding. Sharma et al. (2004) reported the pyramiding of BPH-resistance genes through MAS. However, the PYL with Bph1 and bph2 showed the same level of BPH resistance as a line carrying Bph1 alone owing to a lack of BPH strains that were virulent against Bph1 and bph2. Fujita et al. (2006) demonstrated that the pyramiding of two dominant genes, Grh2 and Grh4, that confer resistance to the green rice leafhopper (GRH, Nephotettix cincticeps Uhler) provided a higher level of antibiosis against a non-selective strain of GRH than a line carrying only one of these two resistance genes. Similarly, three PYLs (which carry Grh2 and Grh6, Grh4 and Grh6, and Grh5 and qGRH4) showed epistatic effects of gene pyramiding (Fujita et al. 2010).
We developed pre-NILs and a pre-PYL carrying BPH25 and BPH26 through MAS using SSR markers tightly linked to the resistance genes by continuous backcrossing with a susceptible japonica cultivar (T65). When we evaluated the current virulence status of BPH against BPH25 and BPH26 using the virulent BPH strain Japan-KG-06, the pre-NILs that carried only BPH25 or BPH26 were susceptible to Japan-KG-06, but the pre-PYL that carried both had a high level of resistance to this strain (Fig. 5). This result was comparable to the one that Myint et al. (2009a) demonstrated previously, in which the pre-NILs and the pre-PYL for BPH25 and BPH26 produced high adult mortality of the Chikugo-89 strain and no development of the female abdomen (Fig. 5). Thus, our results suggest that the combination of BPH25 and BPH26 conferred a high level of resistance to this most recently arrived BPH strain. The pyramiding of resistance genes is obviously an advantageous strategy for increasing the durability of resistance, as it is less likely for insects to simultaneously overcome two or more resistance genes.
References
Cha YS, Ji H, Yun DW, Ahn BO, Lee MC, Suh SC, Lee CS, Ahn EK, Jeon YH, Jin ID, Sohn JK, Koh HJ, Eun MY (2008) Fine mapping of the rice Bph1 gene, which confers resistance to the brown planthopper (Nilaparvata lugens Stål), and development of STS markers for marker-assisted selection. Mol Cells 26:146–151
Chen JW, Wang L, Pang XF, Pan QH (2006) Genetic analysis and fine mapping of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene bph19(t). Mol Gen Genom 275:321–329
Deen R, Ramesh K, Gautam SK, Rao YK, Lakshmi VJ, Viraktamath BC, Brar DS, Ram T (2010) Identification of new gene for BPH resistance introgressed from O. rufipogon. Rice Genet Newsl 25:70–72
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini preparation: version II. Plant Mol Biol Rep 1:19–21
Fujita D, Doi K, Yoshimura A, Yasui H (2006) Molecular mapping of a novel gene, Grh5, conferring resistance to green rice leafhopper (Nephotettix cincticeps Uhler) in rice, Oryza sativa L. Theor Appl Genet 113:567–573
Fujita D, Yoshimura A, Yasui H (2010) Development of near-isogenic lines and pyramided lines carrying resistance genes to green rice leafhopper (Nephotettix cincticeps Uhler) with the Taichung 65 genetic background in rice (Oryza sativa L.). Breed Sci 60:18–27
Gu K, Tian D, Yang F, Wu L, Sreekala C, Wang GT, Yin Z (2004) High-resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor Appl Genet 108:800–807
Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, Kajiya H, Huang N, Yamamoto K, Nagamura Y, Kurata N, Khush GS, Sasaki T (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148:479–494
Hirabayashi H, Ogawa T (1995) RFLP mapping of Bph-1 (Brown planthopper resistance gene) in Rice. Breed Sci 45:369–371
Hirabayashi H, Ideta O, Sato H, Takeuchi Y, Ando I, Nemoto H, Imbe T, Brar DS, Ogawa T (2004) Identification of a resistance gene to brown planthopper derived from Oryza minuta in rice (in Japanese). Breed Res 6(suppl 1):285
IRRI (1975) Annual Report for 1975. International Rice Research Institute, Los Baños
Ishii T, Brar DS, Multani DS, Khush GS (1994) Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa. Genome 37:217–221
Jairin J, Phengrat K, Teangdeerith S, Vanavichit A, Toojinda T (2007a) Mapping of a broad-spectrum brown planthopper resistance gene, Bph3, on rice chromosome 6. Mol Breed 19:35–44
Jairin J, Teangdeerith S, Leelagud P, Phengrat K, Vanavichit A, Toojinda T (2007b) Physical mapping of Bph3, a brown planthopper resistance locus in rice. Maejo Internat J Sci Technol 1:166–177
Jairin J, Sansen K, Wongboon W, Kothcharerk J (2010) Detection of a brown planthopper resistance gene bph4 at the same chromosomal position of Bph3 using two different genetic backgrounds of rice. Breed Sci 60:71–75
Jena KK, Kim SK (2010) Current status of brown planthopper (BPH) resistance and genetics. Rice 3:161–171
Jena KK, Jeung JU, Lee JH, Choi HC, Brar DS (2006) High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t). and marker-assisted selection for BPH resistance in rice (Oryza sativa L.). Theor Appl Genet 112:288–297
Kawaguchi M, Murata K, Ishii T, Takumi S, Mori N, Nakamura C (2001) Assignment of a brown planthopper (Nilaparvata lugens Stål) resistance gene bph4 to the rice chromosome 6. Breed Sci 51:13–18
Khush GS (1979) Genetics of and breeding for the resistance to the brown planthopper. Brown Planthopper: threat to rice production in Asia. International Rice Research Institute (IRRI), Los Baños, pp 321–332
Khush GS, Brar DS (1991) Genetics of resistance to insects in crop plants. Adv Agron 45:223–274
Kim SM, Sohn JK (2005) Identification of a rice gene (Bph1) conferring resistance to brown planthopper (Nilaparvata lugens Stål) using STS markers. Mol Cells 20:30–34
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
McCouch SR, CGSNL (2008) Gene nomenclature system for rice. Rice 1:72–84
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Murai H, Hashimoto Z, Sharma PN, Shimizu T, Murata K, Takumi S, Mori N, Kawasaki S, Nakamura C (2001) Construction of a high-resolution linkage map of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene bph2. Theor Appl Genet 103:526–532
Murata K, Fujiwara M, Kaneda C, Takumi S, Mori N, Nakamura C (1998) RFLP mapping of a brown planthopper (Nilaparvata lugens Stål) resistance gene bph2 of indica rice introgressed into a japonica breeding line ‘Norin PL4’. Genes Genet Syst 73:359–364
Murata K, Fujiwara M, Murai H, Takumi S, Mori N, Nakamura C (2000) Bph9, a dominant brown planthopper resistance gene, is located on the long arm of rice chromosome 12. Rice Genet Newsl 17:84–86
Myint KKM, Matsumura M, Takagi M, Yasui H (2009a) Demographic parameters of long-term laboratory strains of the brown planthopper, Nilaparvata lugens Stål (Homoptera: Delphacidae) on resistance genes, bph20(t) and Bph21(t) in rice. J Fac Agric Kyushu Univ 54:159–164
Myint KKM, Yasui H, Takagi M, Matsumura M (2009b) Virulence of long-term laboratory populations of the brown planthopper, Nilaparvata lugens (Stål), and whitebacked planthopper, Sogatella furcifera (Horváth) (Homoptera: Delphacidae), on rice differential varieties. Appl Entomol Zool 44:149–153
Park DS, Song MY, Park SK, Lee SK, Lee JH, Song SY, Eun MY, Hahn TR, Sohn JK, Yi G, Nam MH, Jeon JS (2008) Molecular tagging of the Bph1 locus for resistance to brown planthopper (Nilaparvata lugens Stål) through representational difference analysis. Mol Genet Genom 280:163–172
Pathak PK, Heinrichs EA (1982) Selection of biotype populations 2 and 3 of Nilaparvata lugens by exposure to resistant rice varieties. Environ Entomol 11:85–90
Rahman ML, Jiang W, Chu SH, Qiao Y, Ham TH, Woo MO, Lee J, Khanam MS, Chin JH, Jeung JU, Brar DS, Jena KK, Koh HJ (2009) High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor Appl Genet 119:1237–1246
Ram T, Deen R, Gautam SK, Ramesh K, Rao YK, Brar DS (2010) Identification of new genes for brown planthopper resistance in rice introgressed from O. glaberrima and O. minuta. Rice Genet Newsl 25:67–69
SAS Institute Inc. (2003) JMP® User’s Guide, Version 5.1, SAS Institute Inc., Cary, North Carolina
Sharma PN, Ketipearachchi Y, Murata K, Torii A, Takumi S, Mori N, Nakamura C (2002) RFLP/AFLP mapping of a brown planthopper (Nilaparvata lugens Stål) resistance gene Bph1 in rice. Euphytica 129:109–117
Sharma PN, Torii A, Takumi S, Mori N, Nakamura C (2004) Marker-assisted pyramiding of brown planthopper (Nilaparvata lugens Stål) resistance genes Bph1 and Bph2 on rice chromosome 12. Hereditas 140:61–69
Sogawa K (1982) The rice brown planthopper: Feeding physiology and host plant interactions. Ann Rev Entomol 27:49–73
Sogawa K (1992) Rice brown planthopper (BPH) immigrants in Japan change biotype. Internat Rice Res Newsl 17:26–27
Sogawa K, Liu GJ, Shen JH (2003) A review on the hyper-susceptibility of Chinese hybrid rice to insect pests. Chin J Rice Sci 17:23–30
Sonoda T, Yoshimura A, Yasui H (2003) Detection of QTLs for antibiosis to brown planthopper, Nilaparvata lugens Stål, in rice, Oryza sativa L. Rice Genet Newsl 20:83–87
Su CC, Zhai HQ, Wang CM, Sun LH, Wan JM (2006) SSR mapping of brown planthopper resistance gene Bph9 in Kaharamana an Indica rice (Oryza sativa L.). Acta Genetica Sinica 33:262–268
Sun L, Su C, Wang C, Zhai H, Wan J (2005) Mapping of a major resistance gene to the brown planthopper in the rice cultivar Rathu Heenati. Breed Sci 55:391–396
Sun LH, Wang CM, Su CC, Liu YQ, Zhai HQ, Wan JM (2006) Mapping and marker-assisted selection of a brown planthopper resistance gene bph2 in rice (Oryza sativa L.). Acta Genetica Sinica 33:717–723
Tanaka K (2000) A simple method for evaluating the virulence of the brown planthopper. Internat Rice Res Notes 25:18–19
Tanaka K, Matsumura M (2000) Development of virulence to resistant rice varieties in the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae), immigrating into Japan. Appl Entomol Zool 35:529–533
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Velusamy R, Chelliah S, Heinrichs EA, Medrano F (1984) Brown planthopper biotypes in India. Internat Rice Res Notes 9:19
Wang S, Basten CJ, Zeng Z-B (2004) Windows QTL Cartographer 2.0. Department of Statistics, North Carolina State University, Raleigh
Yamazaki Y, Sakaniwa S, Tsuchiya R, Nonomura KI, Kurata N (2010) Oryzabase: an integrated information resource for rice science. Breed Sci 60:544–548
Yang H, Ren X, Weng Q, Zhu L, He G (2002) Molecular mapping and genetics of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene. Hereditas 136:39–43
Yara A, Phi CN, Matsumura M, Yoshimura A, Yasui H (2010) Development of near-isogenic lines for BPH25(t) and BPH26(t), which confer resistance to the brown planthopper, Nilaparvata lugens (Stål.) in indica rice ‘ADR52’. Breed Sci 60:639–647
Acknowledgments
We are grateful to the Biotron Application Center, Kyushu University, for use of the Entomotron in which we conducted our experiments. We also thank the staff members of the Research Group for Insect Pest Management, Kyushu Okinawa Agricultural Research Center, for their technical support. This work was partially supported by a Grant-in-Aid for Scientific Research (2058007) from the Ministry of Education, Culture, Sports, Science, and the Ministry of Agriculture, Forestry and Fishery of Japan (Integrated Research Project for Plants, Insects, and Animals using Genome Technology, QT-4008 and QT-4010, and Genomics for Agricultural innovation, QTL-2001 to H.Y).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Tai.
K. K. M. Myint and D. Fujita contributed equally to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Myint, K.K.M., Fujita, D., Matsumura, M. et al. Mapping and pyramiding of two major genes for resistance to the brown planthopper (Nilaparvata lugens [Stål]) in the rice cultivar ADR52. Theor Appl Genet 124, 495–504 (2012). https://doi.org/10.1007/s00122-011-1723-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1723-4