Abstract
Background
Aedes aegypti is a potential vector of West Nile, Japanese encephalitis, chikungunya, dengue and Zika viruses. Alternative control measurements of the vector are needed to overcome the problems of environmental contamination and chemical resistance. Xenorhabdus and Photorhabdus are symbionts in the intestine of entomopathogenic nematodes (EPNs) Steinernema spp. and Heterorhabditis spp. These bacteria are able to produce a broad range of bioactive compounds including antimicrobial, antiparasitic, cytotoxic and insecticidal compounds. The objectives of this study were to identify Xenorhabdus and Photorhabdus isolated from EPNs in upper northern Thailand and to study their larvicidal activity against Ae. aegypti larvae.
Results
A total of 60 isolates of symbiotic bacteria isolated from EPNs consisted of Xenorhabdus (32 isolates) and Photorhabdus (28 isolates). Based on recA gene sequencing, BLASTN and phylogenetic analysis, 27 isolates of Xenorhabdus were identical and closely related to X. stockiae, 4 isolates were identical to X. miraniensis, and one isolate was identical to X. ehlersii. Twenty-seven isolates of Photorhabdus were closely related to P. luminescens akhurstii and P. luminescens hainanensis, and only one isolate was identical and closely related to P. luminescens laumondii. Xenorhabdus and Photorhabdus were lethal to Ae aegypti larvae. Xenorhabdus ehlersii bMH9.2_TH showed 100% efficiency for killing larvae of both fed and unfed conditions, the highest for control of Ae. aegypti larvae and X. stockiae (bLPA18.4_TH) was likely to be effective in killing Ae. aegypti larvae given the mortality rates above 60% at 72 h and 96 h.
Conclusions
The common species in the study area are X. stockiae, P. luminescens akhurstii, and P. luminescens hainanensis. Three symbiotic associations identified included P. luminescens akhurstii-H. gerrardi, P. luminescens hainanensis-H. gerrardi and X. ehlersii-S. Scarabaei which are new observations of importance to our knowledge of the biodiversity of, and relationships between, EPNs and their symbiotic bacteria. Based on the biological assay, X. ehlersii bMH9.2_TH begins to kill Ae. aegypti larvae within 48 h and has the most potential as a pathogen to the larvae. These data indicate that X. ehlersii may be an alternative biological control agent for Ae. aegypti and other mosquitoes.
Similar content being viewed by others
Background
Dengue fever (DF) caused by dengue virus is the most important vector-borne disease transmitted by Aedes spp. In southeast Asia, including Thailand, the primary vector for dengue virus is Aedes aegypti and the secondary vector is Ae. albopictus [1]. Dengue fever is a major risk to public health due to the recent spread of the virus worldwide [2, 3]. This disease threatens approximately 390 million people living in over 100 countries [4, 5]. Aedes spp. are not only the vectors of the dengue virus but also of West Nile virus and chikungunya virus [6]. West Nile virus infects more than 2.5 million people causing over 1300 deaths during 1999–2010 [7]. Chikungunya virus normally does not cause death in humans but has been reported in more than 45 countries with epidemic in India [8]. Recently, the Zika virus, a worldwide public health concern, was shown to be transmitted to humans by Ae. aegypti [4]. More than 200,000 human cases infected with the Zika virus have been reported in the Americas. Approximately 3.6 billion people worldwide are living in at-risk areas for Zika virus, dengue virus, chikungunya, and yellow fever virus transmission [9]. The epidemics of Zika virus and dengue virus are the greatest public health threats to the human population worldwide. There are no specific treatments or vaccines currently available to combat infections by these viruses; the only effective approach to prevent infection is to avoid mosquito bites [10]. Therefore, control of the larval and adult mosquitoes is an essential precautionary measure to prevent the disease. To reduce mosquito population density, life span and human contact, control measures against Aedes include elimination of breeding sites, use of chemical control, genetic and biological control [11]. Elimination of breeding places is a simple technique and low-cost method to protect the breeding sites of Aedes. Chemical control (organochlorides, DDT; organophosphates, OP; pyrethroids) is usually considered as the first method for mosquito control. However, repeated use of pesticides leads to development of insecticide-resistant mosquito populations and toxic to humans. Aedes spp. in all regions of the world have developed resistance to DDT [12]. Pyrethroid resistance in mosquitoes was also reported in several countries in Asia [12]. In addition, the insecticide also causes environmental contamination and destruction of non-target organisms. Genetic control including the sterile insect technique (SIT) and rearing of insects carrying a dominant lethal allele (RIDL) are species-specific methods for Ae. albopictus and Ae. aegypti [13, 14]. However, most of the genetic control methods are in the laboratory conditions, and need more consideration in several aspects such as cost, natural condition and environmental risk assessment [11]. Larval control is the most economical method to eradicate Aedes spp. This method can scope on a certain source with or without pesticide applications rather than spraying miles of chemicals for control of the adult stage.
Biological control is an alternative method used for mosquito vectors. Compared to chemical control it is environmentally friendly as well as sustainable because of the slow pace of resistance development against biological control agents. In addition, the continued use of chemicals over a long period of time can induce insecticide resistance in mosquito populations. The biological control agents used against Aedes mosquitoes include Bacillus thuringiensis israelensis or B. sphericus and their toxins and Xenorhabdus/Photorhabdus [15,16,17,18,19], fungi Metarhizium anisopliae and Beauveria bassiana [20,21,22], the protozoan Acanthamoeba polyphaga [23] and the copepod Macrocyclops albidus [24]. It has been suggested that Bacillus thuringiensis, Xenorhabdus and Photorhabdus have potential for the biological control of Aedes mosquitoes. These entomopathogenic bacteria are used in the control of mosquito larvae [17, 25].
Xenorhabdus and Photorhabdus, Gram-negative bacteria of the family Enterobacteriaceae, are symbiotic bacteria with entomopathogenic nematodes (EPNs) of the genera Steinernema and Heterorhabditis, respectively [26]. These bacteria produce many bioactive compounds which demonstrate insecticidal, cytotoxic and anti-microbial activities [27]. Xenorhabdus and Photorhabdus live in the gut of the infective juvenile stages of EPNs. They can enter the insect hosts by the aid of EPNs through a natural orifice such as the mouth and anus or by direct penetration through the skin. The bacteria are released into the blood system of the insects and then multiply, release toxins and antimicrobial compounds which result in insect septicemia and death within 24–48 h [27]. Symbiotically the EPNs benefit from living with the bacteria by eating the bacteria and the remains of the infected insects. The EPNs grow and reproduce, increasing the number of infective insect juvenile EPNs which then leave the host insect cadaver to find a new host.
Xenorhabdus and Photorhabdus have been successfully used to reduce the development of several insect pests in laboratory conditions [28]. The use of P. luminescens mixed with Bacillus thuringiensis kurstaki inhibits the growth of the African cotton leafworm Spodoptera littoralis (Lepidoptera: Noctuidae) [29]. Forst et al. [30] reported that 20 cells of X. nematophilus killed as much as 90% of the larvae of Manduca sexta. The PirAB toxins from P. asymbiotica exhibited potential for reduction the survival of the larvae of Ae. aegypti and Ae. albopictus larvae. An oral dose of X. nematophila and P. luminescens cell suspension is lethal to Aedes larvae even in the absence of the entomopathogenic nematode [25].
The first survey of symbiotic bacteria in Thailand demonstrated that Xenerhabdus and Photorhabdus are found throughout the country with predomination of X. stockiae and P. luminescens [31]. Subsequently it was found that Xenorhabus sp. PB61.4 isolated from EPN from Chaiyaphum Province in the northeast Thailand are able to produce a novel substance (chaiyaphumine) that is toxic to Plasmodium falciparum in vitro [32]. Thus, Thailand is a natural environment for studying Xenorhabdus and Photorhabdus diversity and for identifying their potential bioactive compounds. We previously isolated and identified the EPNs collected from upper northern Thailand [33]. The present study continues our work on the identification of symbiotic bacteria. Our objectives were to identify Xenorhabdus and Photorhabdus isolated from entomopathogenic nematodes in the upper northern regions of Thailand and to study their larvicidal activity against Ae. aegypti larvae in laboratory conditions. Here we report the identification and phylogenetic analysis of symbiotic bacteria based on sequencing of the housekeeping gene (recA) which gave more reliable results for the taxonomy purposes. In addition, phylogenetic analyses of the recA gene showed the potential topology for distinguishing between Xenorhabdus spp. and Photorhabdus spp.
Methods
Isolation and identification of Xenorhabdus and Photorhabdus
Eight provinces located in the upper northern Thailand including Chiang Mai, Chiang Rai, Nan, Phayao, Phrae, Lampang, Lamphun, and Mae Hong Son were selected for soil sampling. Xenorhabdus and Photorhabdus bacteria were isolated from the hemolymph of larval Galleria mellonella (greater wax moth) cadavers infected with EPNs that were previously isolated and identified as described in Vitta et al. [33]. To propagate EPNs and obtain hemolymphs containing the symbiotic bacteria, water containing approximately 300 EPNs (500 μl) was placed onto a sterile Petri dish containing 5 larvae of G. mellonella. The Petri dish was then sealed with parafilm and incubated in the dark at room temperature. The insect larvae were observed daily for 2–3 days. The resulting insect cadavers were then washed with 95–100% ethanol and placed on another sterile Petri dish. The third segment from the mouth parts of the dead G. mellonella larvae were opened using fine sterile forceps to obtain the hemolymph containing Xenorhabdus and Photorhabdus. A drop of hemolymph was streaked on sterile plates of nutrient bromothymol blue-triphenyltetrazolium chloride agar (NBTA) which were then stored in the dark at room temperature. After 4 days of incubation, preliminary identification of these bacteria was performed by observing the colony morphology. The colonies of species of the genus Xenorhabdus are dark blue, convex, umbonated and swarm while those of species of the genus Photorhabdus are dark green, convex and umbonated [31]. A single colony from each isolate was subcultured on the same medium and kept in Luria-Bertani (LB) broth supplemented with 20% glycerol at -80 °C for further species identification and bioassay.
Species identification of Xenorhabdus and Photorhabdus was carried out by polymerase chain reaction (PCR) and analysis of partial recA gene sequences. Genomic DNA from each bacterial isolate extracted using a genomic DNA mini kit (Geneaid Biotech Ltd., New Taipei, Taiwan). PCR primers [34], reagents and PCR amplication conditions used were as described in Thanwisai et al. [31]. The PCR cycles were conducted in an Applied Biosystems thermal cycler (Life Technologies, Carlsbad, CA, USA). The PCR products were purified using a Gel/PCR DNA Fragment Extraction Kit (Geneaid Biotech Ltd., New Taipei, Taiwan) before sequencing at Macrogen Inc., Korea. All sequences were edited using the SeqManII program (DNASTAR inc., Wisconsin, Madison, USA). A BLASTN search was performed to identify Xenorhabdus and Photorhabdus to the species level by finding the similarity of the recA sequences with known sequences in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic analysis of Xenorhabdus and Photorhabdus
Phylogenetic analysis was performed using MEGA version 5.2 [35]. Twenty three recA sequences for Xenorhabdus spp. and 15 sequences for Photorhabdus spp. were downloaded from the NCBI database; Escherichia coli (GenBank: U00096.3) was used as out-group. Multiple sequences were aligned using Clustal W version 5.2 for editing the nucleotide sequences which were trimmed to 508 bp. Maximum likelihood trees were costructed using the Kimura-2 model (1000 bootstrap replicates).
Biological assay
A total of 12 bacterial isolates of Xenorhabdus (8 isolates) and Photorhabdus (4 isolates) were randomly selected from 60 isolates based on different branches in a maximum-likelihood tree.
The eight Xenorhabdus isolates were divided into 3 groups: (i) Xenorhabdus bMH9.2_TH isolate closely related to X. ehlersii; (ii) Xenorhabdus bMH16.4_TH, bMH16.1_TH, bMH4.5_TH isolates closely related to X. miraniensis; (iii) Xenorhabdus bLPA12.2_TH, bLPA18.4_TH, bCR7.3_TH and bPH23.5_TH isolates closely related to X. stockiae. The Photorhabdus isolates were divided into 3 groups: (i) Photorhabdus bPY17.4_TH and bLPO16.2_TH closely related to P. luminescens hainanensis; (ii) Photorhabdus bNA22.1_TH closely related to P. luminescens akhurstii; and (iii) Photorhabdus bMH8.4_TH closely related to P. luminescens laumondii. All selected Xenorhabdus and Photorhabdus isolates (tested group) were separately grown on NBTA. Escherichia coli ATCC®25922 cultured on Tryptone soy agar (TSA), was used as one negative control and distilled water was used as a second negative control.
A single colony of each isolate of Photorhabdus and Xenorhabdus on NBTA was transferred, in sterile conditions, into a 15 ml tube containing 5 ml of 5YS broth medium which was composed of 5% yeast extract (w/v), 0.5% NaCl (w/v), 0.05% K2HPO4 (w/v), 0.05% NH2H2PO4 (w/v), 0.02% MgSO4.7H2O (w/v) [36]. The tubes were then incubated while shaking (150 rpm) at room temperature for 48 h. Each tube of the bacterial suspension was then centrifuged at 10,000 rpm for 10 min to obtain a bacterial pellet which was then resuspended in sterile distilled water. Adjustment of the bacterial suspension at OD600 to 1.0 was performed by spectrophotometer (BECMAN-COUTER Model DU®730, Fullerton, USA). A single colony of E. coli ATCC®25922 on TSA was subcultured on 5YS broth and then processed under the same conditions applied for Photorhabdus and Xenorhabdus. A 10-fold serial dilution spread plate was performed and the concentration of bacterial suspension was found to be 108 (CFU/ml).
Aedes aegypti third- and fourth-instar larvae were purchased from the Medical Entomology Division of the National Institute of Health, Department of Medical Sciences, Ministry of Public Health of Thailand. The larvae were transported to the Department of Microbiology and Parasitology at the Faculty of Medical Science, Naresuan University. Aedes aegypti larvae were rested in dechlorinated water for one day prior to test.
A biological assay was performed under two conditions; fed condition (larvae exposed to symbiotic bacteria were fed with ground pet food) and unfed condition (larvae exposed to symbiotic bacteria were not given food). Escherichia coli ATCC®25922 and distilled water were used as negative controls. For each bioassay, 30 larvae (10 larvae/well) were transferred to 3 wells of a 24-well microtiter plate. Two ml bacterial suspension of each isolate containing 108 CFU/ml was added in to each well. The plates were kept at room temperature, and the larval mortality was observed daily for 4 days (24, 48, 72 and 96 h). Larvae that showed no movement response after teasing with a fine sterile toothpick were considered dead. All assays were carried out 3 times.
Statistical analysis
The mortality rate (percentages) of the larvae in the control groups and in the treatment group (each bacterial isolate) were statistically tested using SPSS version 17 (Fisher’s exact test; P < 0.05). Fisher’s exact test was performed to determine the difference in mortality rates between the fed and unfed groups.
Results
Identification of Xenorhabdus and Photorhabdus
Based on the colony morphology on the NBTA, 32 isolates of Xenorhabdus were isolated from the EPNs and were preliminarily characterized based on a dark blue, covex and umbonated or swarm colony, while colonies of Photorhabdus (28 isolates) were dark green, convex and umbonated.
A partial region of the recA gene from both Xenorhabdus and Photorhabdus was determined by PCR and sequenced. PCR amplicons of the recA gene of Xenorhabdus and Photorhabdus were approximately 900 bp in size. After BLASTN search, 27 isolates of Xenorhabdus (GenBank: KY404017–KY404048) showed sequence similarity to X. stockiae (97–99% similarity), while 4 isolates to X. miraniensis (98–100% similarity) and only one isolate of Xenorhabdus was recognized as X. ehlersii with 97% similarity. For Photorhabdus (GenBank: KY436924–KY436951), 18 isolates, 9 isolates and 1 isolate showed sequence similarity to P. luminescens akhurstii (98–100% similarity), P. luminescens hainanensis (98–99% similarity), and P. luminescens laumondii (98% similarity), respectively.
Phylogenetic tree of Xenorhabdus and Photorhabdus
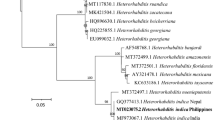
Based on the maximum-likelihood tree based on 508 bp of the recA region, most Xenorhabdus isolates (27 isolates) in Group 1 were closely related to the X. stockiae strain TH01 (GenBank: FJ823425.1). All identified isolates of X. stockiae were associated with Steinernema websteri. Four isolates of Xenorhabdus (Group 2) grouped with X. miraniensis (GenBank: FJ823414.1). All identified isolates of X. miraniensis were hosted by Steinernema websteri. The remaining Xenorhabdus isolate (Group 3) was clustered in X. ehlersii (GenBank: FJ823398.1). Unlike the other two clusters, this Xenorhabdus isolate was associated with Steinernema scarabaei (Fig. 1). From the phylogenetic analysis of Photorhabdus, two main groups (Group 1 and 2) containing most of the isolates of Photorhabdus (27 isolates) were clustered into a group containing P. luminescens akhurstii (GenBank: EU862005.1) and P. luminescens hainanensis (GenBank: EU930342.1) (Fig. 2). Most P. luminescens akhurstii isolates (8 isolates) were associated with Heterorhabditis indica. One isolate each of P. luminescens akhurstii (bCM17.3_TH) and P. luminescens hainanensis (bLPO13.3_TH) were hosted by Heterorhabditis gerrardi. The group with only one isolate of Photorhabdus (Group 3) was grouped together with P. luminescens laumondii (GenBank: FJ861999.1) which was associated with Heterorhabditis sp. SGmg3 (Fig. 2).
Phylogenetic relationships of Xenorhabdus (n = 32) isolated from EPNs of upper northern Thailand and Xenorhabdus spp. worldwide based on maximum-likelihood analysis of 508 bp of the recA region. Bootstrap values are based on 1000 pseudoreplicates. Codes for bacterial isolates indicated in the phylogenetic tree are defined as province/soil-site collection/country, e.g. bMH9.2_TH (b, bacteria; MH, Mae Hong Son Province; 9.2, soil collection site; TH, Thailand). Eight provinces located in the upper northern Thailand including Chiang Mai (CM), Chiang Rai (CR), Nan (NA), Phayao (PY), Phrae (PH), Lampang (LPA), Lamphun (LPO), and Mae Hong Son (MH) were selected for soil sampling
Phylogenetic relationships of Photorhabdus (n = 28) isolated from EPNs of upper northern Thailand and Photorhabdus spp. worldwide based on maximum-likelihood analysis of 508 bp of the recA region. Bootstrap values are based on 1000 pseudoreplicates. Codes for bacterial isolates indicated in the phylogenetic tree are defined as province/soil-site collection/country, e.g. bMH9.2_TH (b, bacteria; MH, Mae Hong Son Province; 9.2, soil collection site; TH, Thailand). Eight provinces located in the upper northern Thailand including Chiang Mai (CM), Chiang Rai (CR), Nan (NA), Phayao (PY), Phrae (PH), Lampang (LPA), Lamphun (LPO), and Mae Hong Son (MH) were selected for soil sampling
Larvicidal activity of Xenorhabdus and Photorhabdus against Aedes aegypti
Xenorhabdus ehlersii (bMH9.2_TH) showed highest effectiveness for killing Aedes aegypti larvae under both fed and unfed conditions. The mortality rates of Ae. aegypti larvae in fed condition were 39%, 96%, 98%, and 100% after exposure to a suspension of X. ehlersii (bMH9.2 TH) for 24, 48, 72 and 96 h, respectively, while the mortality rates of unfed larvae were 56%, 98%, 99% and 100% at similar time intervals (Table 1). These rates were significantly different (Fisher’s exact test, P < 0.05) when compared with the mortality rates of Ae. aegypti larvae in the control groups which were 1% and 2% for Escherichia coli ATCC®25922 and distilled water in fed condition, and 7% and 0% for E. coli ATCC® 25,922 and distilled water in unfed condition (Table 1). Xenorhabdus stockiae (bLPA18.4_TH) is likely to be effective in killing Ae. aegypti larvae given the mortality rates above 60% at 72 and 96 h under both fed and unfed condition. However, the other isolates of X. stockiae (bLPA12.2_TH, bCR7.3_TH and bPH23.5_TH) were less pathogenic to the larvae, exhibiting a mortality rates lower than 50%. All isolates of X. miraniensis (bMH16.4_TH, bMH16.1_TH and bMH4.5_TH) showed a negligible toxicity to Ae. aegypti larvae and the observed mortality rate was lower than 20%. All Photorhabdus isolates (bPY17.4_TH, bLPO16.2_TH, bMH8.4_TH and bNA22.1_TH) showed low pathogenic effect on Ae. aegypti larvae upon oral uptake, with a mortality rate lower than 40% (Table 1). The mortality rates were significantly different between fed and unfed mosquito larvae in case of P. luminescens hainanensis (bLPO16.2_TH) and X. stockiae (bCR7.3_TH and bPH23.5_TH) (see Table 2).
Discussion
Xenorhabdus stockiae and P. luminescens akhurstii were the most common symbiotic bacteria that we found in EPNs of upper northern Thailand. Most X. stockiae isolates were associated with S. websteri while P. luminescens akhurstii was associated with H. indica [33]. This is consistent with a previous report showing that 66 isolates of X. stockiae and three isolates of X. miraniensis were isolated from S. websteri and 57 isolates of P. luminescens hainanensis and a few isolates of P. luminescens laumondii and P. luminescens akhurstii were associated with H. indica [31]. This suggests that the common association in the study area is X. stockiae - S. websteri and P. luminescens akhurstii - H. indica.
Two Photorhabdus isolates, P. luminescens akhurstii (bCM17.3_TH) and P. luminescens hainanensis (bLPO13.3_TH), were associated with Heterorhabditis gerrardi. Photorhabdus luminescens akhurstii was previously reported to be in a symbiotic association with H. indica [34]. This suggests that P. luminescens akhurstii can be hosted by not only H. indica but also by H. gerrardi. The EPN H. gerrardi was previously reported as being associated with P. asymbiotica [37]. Photorhabdus luminescens akhurstii and H. gerrardi is a new EPN-symbiotic bacteria association. Photorhabdus luminescens hainanensis were hosted by Heterorhabditis sp. in China [34]. To our knowledge, this is the first record of the association between P. luminescens hainanensis and H. gerrardi. In our study, one isolate of X. ehlersii was shown to be associated with S. scarabaei. Previous studies reported that X. ehlersii is associated with S. serratum [38] and S. longicaudum [39] while S. sarabaei is associated with X. koppenhoeferi [40]. The symbiotic relationship of X. ehlersii - S. scarabaei complex is a novel association observed and reported in our study.
Phylogenetic analysis of 32 isolates of Xenorhabdus showed that most Xenorhabdus isolates are closely related to X. stockiae which is similar to previous reports [31]. A small number of Xenorhabdus isolates were closely related to X. miraniensis and only one isolate was closely related to X. ehlersii isolated from China [38]. In our study we found X. stockiae as the majority of the isolates of the genus Xenorhabdus. Most of the Photorhabdus isolates were closely related to P. luminescens akhurstii and P. luminescens hainanensis. P. luminescens akhurstii was predominant among the isolates of the genus Photorhabdus. Three new associations between EPN and symbiotic bacteria which were observed in this study: H. gerrardi - P. luminescens akhurstii, H. gerrardi - P. luminescens hainanensis and S. scarabaei - X. ehlersii complexes.
Xenorhabdus ehlersii bMH9.2_TH killed up to 100% of Ae. aegypti at 96 h, in both fed and unfed conditions. Previous studies on the same group of symbiotic bacteria reported that a cell suspension of P. luminescens kills up to 73% of Ae. aegypti larvae in fed groups, and 83% in unfed groups, followed by X. nematophila with larval mortality of up to 52% in the fed condition and 42% in the unfed condition [25]. This may be due to X. ehlersii bMH9.2_TH producing bioactive compounds that are effective in killing mosquito larvae. XeGroEL protein produced from X. ehlersii has been proven to kill Galleria mellonella [41, 42]. In addition, other species of Xenorhabdus such as X. nematophila can produce compounds with insecticidal properties such as toxin complexes or lipopolysaccharides [43,44,45]. All compounds mentioned above were suggested to play roles as insecticidal compounds. In addition, X. stockiae (bLPA18.4_TH) in this study was considered an effective entomopathogen to kill Ae. aegypti larvae. Xenorhabdus stockiae PB09 was previously reported to have acaricidal and antibacterial activities [46, 47].
Thus our study is the first report of insecticidal activity of X. stockiae. This suggests that X. stockiae may produce several bioactive compounds as it manifests several bioactivities: however, there are no reports on purified bioactive compounds from X. stockiae. Bioactive compounds from X. stockiae isolates should be further investigated for better understanding of their mechanism of action. Unlike X. stockiae (bLPA18.4_TH), other X. stockiae isolates (bLPA12.2_TH, bCR7.3_TH and bPH23.5_TH) showed lower pathogenicity against Ae. aegypti larvae as indicated by the low mortality rates. This may be due to differences in the ability to produce bioactive compounds among different X. stockiae isolates. In this study, we cannot conclude that any substance resulted in the death of Ae. aegypti larvae, but Xenorhabdus and Photorhabdus showed larvicidal activity by oral toxicity against Ae. aegypti. In addition, the mechanisms of Xenorhabdus and Photorhabdus for killing Ae. aegypti larvae are difficult to explain. The isolation of compounds produced by X. ehlersii bMH9.2_TH and X. stockiae bLPA18.4_TH therefore also need further investigation for a better understanding of the mode of action of the bioactive compounds.
The advantage of the use of the bacteria studied here is rapidly killing Aedes spp. larvae within 48 h; these also may be non-toxic to humans. Comparing to other bio-larvicides, Aedes spp. may develop resistance to Beauveria bassiana, Metarhizium anisopliae and Wolbachia [48,49,50]. Aedes spp., can escape copepod predation [51, 52]. Bacillus thuringiensis is more effective on the first and second larval stage than on the third and fourth larval stage of Aedes spp. but Xenorhabdus and Photorhabdus may be effective on all stages of Aedes larvae.
Conclusions
In summary, 60 isolates of symbiotic bacteria from EPNs from upper northern Thailand consisted of 32 Xenorhabdus and 28 Photorhabdus isolates. The common species in the study area were X. stockiae, P. luminescens akhurstii and P. luminescens hainanensis while lower numbers of isolates of X. miraniensis, X. ehlersii and P. luminescens laumondii were observed. Three novel symbiotic associations identified included P. luminescens akhurstii - H. gerrardi, P. luminescens hainanensis - H. gerrardi and X. ehlersii - S. scarabaei which are of importance to our knowledge of the biodiversity of, and relationships between, EPNs and their symbiotic bacteria. Based on our bioassay, Xenorhabdus and Photorhabdus isolates are lethal to Ae. aegypti larvae. Different species and strains of symbiotic bacteria caused different levels of mortality in Ae. aegypti. Xenorhabdus ehlersii bMH9.2_TH begins to kill Ae. aegypti larvae within 48 h and is the best pathogen to the larvae. This indicates that X. ehlersii may be an alternative biological control agent for Ae. aegypti and other mosquitoes. The bioactive compounds in X. ehlersii should be investigated for further understanding of the application of these bacteria and their bioactive derivatives to the bio-control of mosquitoes.
Abbreviations
- CFU/ml:
-
Colony forming unit/ml
- DDT:
-
Dichlorodiphenyltrichloroethane
- DF:
-
Dengue fever
- EPN:
-
Entomopathogenic nematode
- LB:
-
Luria-Bertani
- NBTA:
-
Nutrient bromothymol blue -triphenyltetrazolium chloride agar
- OP:
-
Organophosphates
- PirAB:
-
Photorhabdus insect related AB
- recA:
-
recombinase A
- TSA:
-
Tryptone soy agar
References
Vythilingam I, Sam JI, Chan YF, Khaw LT, Sulaiman YW. New paradigms for virus detection, surveillance and control of Zika virus vectors in the settings of Southeast Asia. Front Microbiol. 2016;7:1452.
Laughlin CA, Morens DM, Cassetti MC, Costero-Saint Denis A, San Martin JL, Whitehead SS, Fauci AS. Dengue research opportunities in the Americas. J Infect Dis. 2012;206:1121–7.
Mahesh Kumar P, Kovendan K, Murugan K. Integration of botanical and bacterial insecticide against Aedes aegypti and Anopheles stephensi. Parasitol Res. 2013;112:761–71.
Benelli G, Mehlhorn H. Declining malaria, rising of dengue and Zika virus: insights for mosquito vector control. Parasitol Res. 2016;115:1747–54.
Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyer CL, et al. The global distribution and burden of dengue. Nature. 2013;496:504–7.
Mint Mohamed Lemine A, Ould Lemrabott MA, Hasni Ebou M, Mint Lekweiry K, Ould Ahmedou Salem MS, Ould Brahim K, et al. Mosquitoes (Diptera: Culicidae) in Mauritania: a review of their biodiversity, distribution and medical importance. Parasit Vectors. 2017;10(1):35.
Kilpatrick AM. Globalization, land use, and the invasion of West Nile virus. Science. 2011;334:323–7.
Lahariya C, Pradhan SK. Emergence of chikungunya virus in Indian subcontinent after 32 years: a review. J Vector Borne Dis. 2006;43:151–60.
Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. An update on Zika virus infection. Lancet. 2017; doi:10.1016/S0140-6736(17)31450-2.
WHO. Zika virus. Fact sheet: Updated 6 September 2016. 2016. http://www.who.int/mediacentre/factsheets/zika/en/. Accessed 27 Dec 2016.
Baldacchino F, Caputo B, Chandre F, Drago A, Della Torre a, Montarsi F, et al. Control methods against invasive Aedes mosquitoes in Europe: a review. Pest Manag Sci. 2015;71:1471–85.
Naqqash MN, Gokce A, Bakhsh A, Salim M. Insecticide resistance and its molecular basis in urban insect pests. Parasitol Res. 2016;115:1363–73.
Bellini R, Medici A, Puggioli A, Balestrino F, Carrieri M. Pilot field trials with Aedes albopictus irradiated sterile males in Italian urban areas. J Med Entomol. 2013;50:317–25.
Winskill P, Harris AF, Morgan SA, Stevenson J, Raduan N, Alphey L, et al. Genetic control of Aedes aegypti: data-driven modelling to assess the effect of releasing different life stages and the potential for long-term suppression. Parasit Vectors. 2014;7:68.
Kovendan K, Murugan K, Vincent S, Kamalakannan S. Larvicidal efficacy of Jatropha curcas and bacterial insecticide, Bacillus thuringiensis, against lymphatic filarial vector, Culex quinquefasciatus say (Diptera: Culicidae). Parasitol Res. 2011;109:1251–7.
Kovendan K, Murugan K, Vincent S, Barnard DR. Studies on and pupicidal activity of Leucas aspera Willd. (Lamiaceae) and bacterial insecticide, Bacillus sphaericus, against malarial vector, Anopheles stephensi Liston (Diptera: Culicidae). Parasitol Res. 2012;110:195–203.
Park Y. Entomopathogenic bacterium, Xenorhabdus nematophila and Photorhabdus luminescens, enhances Bacillus thuringiensis Cry4Ba toxicity against yellow fever mosquito, Aedes aegypti (Diptera: Culicidae). J Asia Pac Entomol. 2015;18:459–63.
Setha T, Chantha N, Benjamin S, Socheat D. Bacterial larvicide, Bacillus thuringiensis israelensis strain AM 65-52 water dispersible granule formulation impacts both dengue vector, Aedes aegypti (L.) population density and disease transmission in cambodia. PLoS Negl Trop Dis. 2016;10:e0004973.
Park Y, Kyo Jung J, Kim Y. A mixture of Bacillus thuringiensis subsp. israelensis with Xenorhabdus nematophila-cultured broth enhances toxicity against mosquitoes Aedes albopictus and Culex pipiens pallens (Diptera: Culicidae). J Econ Entomol. 2016; doi:https://doi.org/10.1093/jee/tow063.
Leles RN, Alessandro WB, Luz C. Effects of Metarhizium anisopliae conidia mixed with soil against the eggs of Aedes aegypti. Parasitol Res. 2012;110:1579–82.
Gomes SA, Paula AR, Ribeiro A, Moraes CO, Santos JW, Silva CP, et al. Neem oil increases the efficiency of the entomopathogenic fungus Metarhizium anisopliae for the control of Aedes aegypti (Diptera: Culicidae) larvae. Parasit Vectors. 2015;8:669.
Jaber S, Mercier A, Knio K, Brun S, Kambris Z. Isolation of fungi from dead arthropods and identification of a new mosquito natural pathogen. Parasit Vectors. 2016;9:491.
Otta DA, Rott MB, Carlesso AM, da Silva OS. Prevalence of Acanthamoeba spp. (Sarcomastigophora: Acanthamoebidae) in wild populations of Aedes aegypti (Diptera: Culicidae). Parasitol Res. 2012;111:2017–22.
Veronesi R, Carrieri M, Maccagnani B, Maini S, Bellini R. Macrocyclops albidus (Copepoda: Cyclopidae) for the biocontrol of Aedes albopictus and Culex pipiens in Italy. J Am Mosq Control Assoc. 2015;31:32–43.
da Silva OS, Prado GR, Silva CE, Costa M, Heermann R. Oral toxicity of Photorhabdus luminescens and Xenorhabdus nematophila (Enterobacteriaceae) against Aedes aegypti (Diptera: Culicidae). Parasitol Res. 2013;112:2891–6.
Forst S, Dowds B, Boemare NE, Stackebrandt E. Xenorhabdus spp. and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47–72.
Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol. 2009;13:224–30.
Luca R, Alberto S, Ignazio F. Emerging entomopathogenic bacteria for insect pest management. Bull Insectology. 2013;2:181–6.
Touzri DB, Amira AB, Khedher SB, Givaudan A, Jaoua S, Tounsi S. Combinatorial effect of Bacillus thuringiensis kurstaki and Photorhabdus luminescens against Spodoptera littoralis (Lepidoptera: Noctuidae). J Basic Microbiol. 2013;53:1–6.
Ahantarig A, Chantawat N, Waterfield NR, Ffrench-Constant R, Kittayapong P. PirAB toxin from Photorhabdus asymbiotica as a larvicide against dengue vectors. J Appl Environ Microbiol. 2009;75:4627–9.
Thanwisai A, Tandhavanant S, Saiprom N, Waterfield NR, Long PK, Bode HB, et al. Diversity of Xenorhabdus and Photorhabdus spp and their symbiotic entomopathogenic nematodes from Thailand. PLoS One. 2012;7:e43835.
Grundmann F, Kaiser M, Schiell M, Batzer A, Kurz M, Thanwisai A, et al. Antiparasitic Chaiyaphumines from entomopathogenic Xenorhabdus sp. PB61.4. J Nat Prod. 2014;77:779–83.
Vitta A, Fukruksa C, Yimthin T, Deelue K, Sarai C, Polseela R, et al. Preliminary survey of entomopathogenic nematodes in upper northern Thailand. Southeast Asian J Trop Med Public Health. 2017;48:18–26.
Tailliez P, Laroui C, Ginibre N, Paule A, Pages S, Boemare N. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis. Int J Syst Evol Microbiol. 2010;60:1921–37.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Shrestha YK, Lee KY. Oral toxicity of Photorhabdus culture media on gene expression of the adult sweet potato whitefly, Bemisia tabaci. J Invertebr Pathol. 2012;109:91–6.
Plichta KL, Joyce SA, Clarke D, Waterfield N, Stock SP. Heterorhabditis gerrardi n. sp. (Nematoda: Heterorhabditidae): the hidden host of Photorhabdus asymbiotica (Enterobacteriaceae: gamma-Proteobacteria). J Helminthol. 2009;83:309–20.
Lengyel K, Langb E, Fodorc A, Szallasc E, Schumannb E. Description of four novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus budapestensis sp. nov., Xenorhabdus ehlersii sp. nov., Xenorhabdus innexi sp. nov., and Xenorhabdus szentirmaii sp. nov. Syst Appl Microbiol. 2005;28:115–22.
Tailliez P, Pagès S, Ginibre N, Boemare N. New insight into diversity in the genus Xenorhabdus, including the description of ten new species. Int J Syst Evol Microbiol. 2006;56:2805–18.
An R, Grewal PS. Comparative analysis of Xenorhabdus koppenhoeferi gene expression during symbiotic persistence in the host nematode. PLoS One. 2016;11:e0145739.
Shi H, Zeng H, Yang X, Zhao J, Chen M, Qiu D. An insecticidal protein from Xenorhabdus ehlersii triggers prophenoloxidase activation and hemocyte decrease in Galleria mellonella. Curr Microbiol. 2012;64:604–10.
Shi H, Zeng H, Yang X, Liu Z, Qiu D. An insecticidal protein from Xenorhabdus ehlersii stimulates the innate immune response in Galleria mellonella. World J Microbiol Biotechnol. 2013;29:1705–11.
Owuama CI. Entomopathogenic symbiotic bacteria, Xenorhabdus and Photorhabdus of nematodes. World J Microbiol Biotechnol. 2001;17:505–15.
Fferench-constant RH, Dowling A, Waterfield NR. Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon. 2007;49:436–51.
Hinchliffe SJ, Hares MC, Dowwling AJ, Ffrench-Constant RH. Insecticidal toxin from the Photorhabdus and Xenorhabdus bacteria. Open Toxinology J. 2010;3:101–18.
Bussaman P, Sa-Uth C, Rattanasena P, Chandrapatya A. Acaricidal activities of whole cell suspension, cell-free supernatant, and crude cell extract of Xenorhabdus stokiae against mushroom mite (Luciaphorus sp.). J Zhejiang Univ Sci B. 2012;13:261–6.
Bussaman P, Rattanasena P. Additional property of Xenorhabdus stockiae for inhibiting cow mastitis- causing bacteria. Biosci Biotech Res Asia. 2016;13:1871–8.
Darbro JM, Johnson PH, Thomas MB, Ritchie SA, Kay BH, Ryan PA. Effects of Beauveria bassiana on survival, blood-feeding success, and fecundity of Aedes aegypti in laboratory and simi-field conditions. Am J Trop Med Hyg. 2012;86:656–64.
Scholte EJ, Takken W, Knols BG. Infection of adult Aedes aegypti and Ae. albopictus mosquitoes with the entomopathogenic fungus Metarhizium anisopliae. Acta Trop. 2007;102:151–8.
Calvitti M, Moretti R, Skidmore AR, Dobson SL. Wolbachia strain wPip yields a pattern of cytoplasmic incompatibility enhancing a Wolbachia-based suppression strategy against the disease vector Aedes albopictus. Parasit Vectors. 2012;5:254.
Marten GG, Reid JW. Cyclopoid copepods. J Am Mosq Control Assoc. 2007;23(Suppl. 2):65–92.
Marten GG. Evaluation of cyclopoid copepods for Aedes albopictus control in tires. J Am Mosq Control Assoc. 1990;6:681–8.
Acknowledgements
We would like to thank Naresuan University for financial support (Grant Number R2556B060, R2559C190 and R2560B055). We would also like to thank Miss Nisarat Chonklin, Miss Sudarat Inyoung, Miss Khanittha Phumdoung, Miss Nongluck Lertjarukkulthaworn, Mr. Repee Thummepak, Mr. Waranun Yotpunya and Mr. Kitsakorn Deelue for soil collections. Many thanks are extended to Mr. Roy Morien of the Naresuan University Language Centre (NULC) for his editing assistance and advice on English.
Funding
This study was supported by Naresuan University (Grant Number R2556B060, and Grant Number R2560B055 for AV and Grant Number R2559C190 for AT).
Availability of data and materials
All data generated or analysed during this study are included in this published article. The seqiences were submitted to the GenBank database under the accession numbers KY404017–KY404048 (Xenorhabdus) and KY436924–KY436951 (Photorhabdus).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: AV and AT. Performed the experiments: CF, TY, MS, AV and PM. Analyzed the data: AT, AV, ST and CF. Contributed reagents/materials/analysis tools: AT and AV. Wrote the paper: AV, CF, AT and ST. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Fukruksa, C., Yimthin, T., Suwannaroj, M. et al. Isolation and identification of Xenorhabdus and Photorhabdus bacteria associated with entomopathogenic nematodes and their larvicidal activity against Aedes aegypti . Parasites Vectors 10, 440 (2017). https://doi.org/10.1186/s13071-017-2383-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-017-2383-2