Abstract
Background
Entomopathogenic nematodes (EPNs) harboring symbiotic bacteria are one of the safest alternatives to the chemical insecticides for the control of various insect pests. Infective juveniles of EPNs locate a target insect, enter through the openings, and reach the hemocoel, where they release the symbiotic bacteria and the target gets killed by the virulence factors of the bacteria. Photorhabdus with Heterorhabditis spp. are well documented; little is known about the associated bacteria.
Main body
In this study, we explored the presence of symbiotic and associated bacteria from Heterorhabditis sp. (IISR-EPN 09) and characterized by phenotypic, biochemical, and molecular approaches. Six bacterial isolates, belonging to four different genera, were recovered and identified as follows: Photorhabdus luminescens, one each strain of Providencia vermicola, Pseudomonas entomophila, Alcaligenes aquatilis, and two strains of Alcaligenes faecalis based on the phenotypic, biochemical criteria and the sequencing of 16S rRNA gene.
Conclusion
P. luminescens is symbiotically associated with Heterorhabditis sp. (IISR-EPN 09), whereas P. vermicola, P. entomophila, A. aquatilis, and A. faecalis are the associated bacteria. Further studies are needed to determine the exact role of the bacterial associates with the Heterorhabditis sp.
Similar content being viewed by others
Background
Entomopathogenic nematodes (EPNs) are obligate pathogens of insects in nature. The infective juveniles (IJs) of EPNs harbor the bacterial symbionts in their gut, and after entry into the target insect, the bacterial symbionts are released into the insect hemocoel. The IJs feed on the insect hemolymph and multiply enormously; the host dies within 24–72 h due to synergistic action of the nematode and the symbiotic bacteria (Ali et al. 2005). Both the nematode and the bacteria go on multiplying on the insect cadaver until a new generation of IJs emerges, which further carry the symbionts to another target host and the infection cycle continues (Kaya and Gaugler 1993). EPNs from the Heterorhabditis and Steinernema genera are symbiotically associated with bacteria from genus Photorhabdus and Xenorhabdus, respectively. Photorhabdus is found throughout the whole intestine whereas Xenorhabdus is sheltered inside a special vesicle in an interior part of intestine (Bird and Akhurst 1983). In the absence of their symbionts, nematodes cannot develop beyond the J1 stage. It is due to the combined effect of both the symbionts that the insect pest is killed in a shorter period of time. EPNs due to their wide host range, ease at handling, short life cycle, amenability to large scale production, and eco-friendly nature have a great potential to be used as biological control agents against a wide range of insect pests (Ali et al. 2005; Dillman and Sternberg 2012).
The ease of their production and exemption from registration requirements are the two big reasons for commercial developments of EPNs. They are widely distributed in different soil habitats and are diverse with respect to the conditions needed for their survival, infectivity potential, mode of replication, and host range (Ali et al. 2005).Other major bacterial agents used in biological control of insect pests are Bacillus thuringiensis and B. sphaericus; these bacteria have been used in controlling insect pests belonging to the orders Diptera, Lepidoptera, and Coleoptera (Vega and Kaya 2012). Invariably, this group of bacteria is only effective after a susceptible insect has ingested their toxin (Pigott and Ellar 2007). This type of “passive infection” apparently limits the degree of efficiency of these bacteria to control the pests. However, the symbiotic bacteria of EPNs (Photorhabdus and Xenorhabdus spp.) are capable of causing “active infection,” which have a wide host range and are introduced into an insect host by their EPNs symbiont. Their pathogenicity is based on their ability to multiply within the hemocoel, elaborate toxic substances, and cause septicemia. The nematode bacteria complex is presently used for controlling a variety of pests in agriculture (Ruiu 2015).
Previously, some exploratory studies have been carried out to isolate EPNs from spice agro-ecosystems and to evaluate them against insect pests (Pervez et al. 2014a). However, knowledge of their symbiotic and associated bacteria is lacking. Although the interaction of Photorhabdus and Xenorhabdus with Heterorhabditis spp. and Steinernema spp., respectively, are well documented, little is known about the associated bacteria. However, it is increasingly apparent that other bacteria co-inhabit or colonize the insect cadaver and the nematode in addition to the typical symbionts.
Hence, present study aimed to characterize the symbiotic and associated bacteria from Heterorhabditis sp. (IISR-EPN 09), using polyphasic approach.
Main text
Materials and methods
Entomopathogenic nematode and insect sources
Infective juveniles (IJs) of Heterorhabditis sp. (IISR-EPN 09) were cultured as per the procedure described by Woodring and Kaya (1988). Freshly harvested IJs were surface sterilized with 0.01% hyamine solution and stored in distilled water in tissue culture flasks for further study. Greater wax moth Galleria mellonella (L.) larvae were reared on artificial diet as per the procedure described by David and Kurup (1988). The larvae were sorted out visually and those of the same size (18 mm) were taken for the study.
Isolation of bacteria
The symbiotic and the associated bacteria from Heterorhabditis sp. (IISR-EPN 09) were isolated by the following 3 methods; in all the 3 methods, the freshly harvested IJs of Heterorhabditis sp. (IISR-EPN 09) were collected and sterilized in 0.01% hyamine solution for 2 min and washed thrice in sterile distilled water. The last wash was plated as control to check the efficacy of the surface sterilization procedure.
Maceration method
The IJs were punctured by using a sterile platinum wire (Akhurst and Boemare 1988). The suspension containing the body contents was diluted and plated on nutrient agar supplemented with 25 mg l−1 bromothymol blue and 40 mg l−1 2,3,5ά triphenyltetrazolium chloride (NBTA) indicator plates (Akhurst 1980). Bacterial colonies were picked and purified subsequently on nutrient agar plates maintained at 28 °C for 72 h. The stock cultures were maintained at −80 °C.
Incubation method
Surface sterilized IJs were directly inoculated into microfuge tubes containing 1 ml nutrient broth. The tubes were incubated at 28 °C for 72 h. After observing turbidity in the medium, a loop full was streaked on to NBTA plates and purified subsequently on nutrient agar.
Hemolymph drop method
G. mellonella larvae infected with Heterorhabditis sp. (IISR-EPN 09) were surface sterilized in 70% ethanol and dissected by using a sterile scalpel under a laminar air flow. A drop of hemolymph was collected from the infected larvae, streaked onto NBTA medium, and incubated at 28 °C for 72 h (Park et al. 1999). Following incubation, the colonies were picked and subsequently purified on nutrient agar.
Phenotypic characterization
The growth characteristics of each isolate were studied at 28 °C and pH 7.0. The colony characteristics like size, consistency, pigmentation, color, form, and margins were recorded on nutrient agar supplemented with 2,3,5-triphenyltetrazolium chloride (TTC). The isolates were further subjected to Gram’s reaction as described previously (Smibert and Krieg 1994).
Biochemical characterization
The biochemical tests to characterize the bacterial isolates viz., catalase, oxidase, indole, methyl red, Voges-Proskauer, citrate utilization, urease, triple sugar iron (TSI), esculin hydrolysis, lipase production, carbohydrate fermentation test using sugars glucose, lactose, sucrose, maltose, and mannitol were carried out using standard protocols.
Amplification and sequencing of 16S rRNA gene
The genomic DNA from the bacterial isolates was extracted from 10 ml of overnight grown cultures and purified manually, using phenol chloroform isoamyl alcohol method. The cells were lysed by using lysozyme and proteinase K as described by Sambrook and Russell (2001). The quality and quantity of the genomic DNA was assessed by agarose gel electrophoresis and spectrophotometrically by using Biophotometer (Eppendorf, Germany). One hundred nanograms of the purified DNA from each isolate was used for the amplification of 16S rRNA gene in a 50-μl reaction volume. The universal primers used for the amplification were 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAGCCGCA-3′) (Zhou et al. 1997). The mixture further contained 1X Taq buffer, MgCl2 at 1.5 mM, dNTP mix at 200 mM, sense and antisense primers at 10 pM each, and 1 U of Taq DNA polymerase. The conditions for the amplification were as: initial denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 60 s, annealing at 52 °C 30 s, extension at 72 °C for 90 s, and a final extension at 72 °C for 5 min. The amplified products were agarose electrophoresed and visualized under UV, followed by gel purification of the desired amplicon using GeneJET gel extraction kit (Fermentas, USA) to remove the unutilized dNTPs and primers. The purified amplicons were sequenced using the same primers that were used for the amplification with fluorescent terminators (Big Dye, Applied Bio systems; Eurofins Genomics India). The identity of the sequences acquired was ascertained by performing BLAST (Altschul et al. 1990).
Phylogenetic analysis
The evolutionary relationship of the 16S rRNA gene sequences was inferred, using the neighbor-joining method (Saitou and Nei 1987) in MEGA v 6.0 (Tamura et al. 2013). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein 1985). The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2013) and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated.
Results and discussion
Isolation of the bacteria
The absence of growth in the plates inoculated with the last wash after surface sterilization confirmed that the surface sterilization procedure using hyamine (0.01%) solution for 2 min was effective. Bacteria were successfully isolated on NBTA (Supplementary Fig. 1) from the gut of EPNs, using all the 3 methods, but rate of recovery varied among the methods. A total of 6 isolates showing distinct and characteristic colony characteristics were taken for further studies (Table 1).
Phenotypic characterization
The colony characteristics like size, shape, elevation, texture, and pigmentation of each isolate was studied and recorded after 24 h of growth at 28 °C (Table 2). All the 6 isolates were found to be rod shaped, motile, and gave negative reaction to Gram’s stain (Table 2; Fig. 1). Among the bacterial isolates, 5 had regular colony shape and all 6 displayed smooth texture. All the isolated bacteria showed red or dark red color colonies with off white marginson TTC (Table 2).
Biochemical characterization
The results of the biochemical tests performed for all the 6 bacterial isolates recovered from Heterorhabditis sp. (IISR-EPN 09) are given in (Table 3 and Fig. 2). All the isolates were positive for the production of catalase, ability to utilize glucose, and ability to utilize citrate as sole source of carbon, whereas the isolates were unable to utilize lactose, produce urease, and hydrolyze esculin. All the 6 isolates, except IISR-EPN BC 06, were able to ferment maltose and 2 out of 6 isolates (IISR-EPN BC 06 and IISR-EPN BC 10) were unable to ferment mannitol (Table 3).
Biochemical characterization of the bacterial isolates. a Indole production, b methyl red (MR), c Voges-Proskauer (VP), d citrate utilization, e urease production, f triple sugar iron (TSI) utilization, and g esculin hydrolysis; fermentation of h glucose, i lactose, j sucrose, k maltose, and l mannitol
Molecular characterization
Single discrete bands of approximately 1500 bp in size corresponding to the 16S rRNA gene were successfully obtained in all the six isolates. The amplicons were sequenced, and the sequences obtained were trimmed and subjected to BLAST. Based on the BLAST analysis, the isolates showed identity with different species of gram-negative bacteria belonging to family Enterobacteriaceae (Table 1). The use of 16S rRNA sequencing for bacterial identification is a reliable method and when used in combination with other classical methods (morphological and biochemical) its accuracy increases further. The various reasons 16S rRNA are used for reliable identification of prokaryotes are as follows: its widespread presence in all bacteria and Achaea, it has both conserved and variable domains (helping in tracing the evolutionary changes over a period of time), and its length (≈ 1500 bp) is ideal for sequencing and analysis (Patel 2001).
Of the 3 methods used for isolation of symbiotic bacteria, 2 methods yielded Alcaligenes faecalis and one led to the recovery of Photorhabdus luminescens, Alcaligenes aquatilis, Pseudomonas entomophila, and Providencia vermicola (Table 1). The actual symbiont P. luminescens was obtained only by hemolymph drop method; however, it would be difficult to establish the supremacy of any of the 3 methods owing to the small sample size taken and the limited number of isolates recovered. The description of the isolates identified is as the following.
Providencia vermicola (IISR-EPN-BC 06)
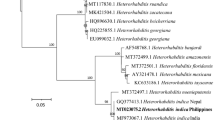
Providencia vermicola are gram-negative rods. Colonies are circular, shining, slimy, convex, and opaque with a brownish center and hyaline periphery. IISR-EPN-BC 06 showed 99% similarity with P. vermicola strain 6G (accession no. KC775772), P. vermicola strain FFA6 (accession no. JN092794), and P. vermicola strain CGS6 (accession no. KF886276). The optimal tree with the sum of branch length was 18.19960570. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 13 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1170 positions in the final dataset (Fig. 3 a).
16S rRNA gene sequence-based phylogenetic relationships of the associated and symbiotic bacterial isolates, (a) Providencia vermicola strain IISR-EPN BC 06, (b) Alcaligenes aquatilis strain IISR-EPN BC 07, (c) A. faecalis strain IISR-EPN BC 08, (d) Pseudomonas entomophila strain IISR-EPN BC 10, and (e) Photorhabdus luminescens strain IISR-EPN BC 12. The values at the branches represent bootstrap values (mean of 1000 repetitions) calculated by neighbor-joining method
Alcaligenes aquatilis (IISR-EPN BC 07)
Alcaligenes aquatilis is a gram-negative, rod-shaped, motile, non-nitrate reducing, oxidase positive, catalase positive, alpha hemolytic, and citrate-positive obligate aerobe that is commonly found in the environment. A. aquatilis belongs to phylum Proteobacteria, class Betaproteobacteria, order Burkholderiales, and family Alcaligenaceae. IISR-EPN BC 07 shows 97% similarity with A. aquatilis strain LMG22996 (accession no. NR104977). The optimal tree with the sum of branch length was 12.36487468. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 12 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1204 positions in the final dataset (Fig. 3 b).
Alcaligenes faecalis (IISR-EPN BC 08 and 11)
A. faecalis is a gram-negative, rod-shaped, motile, non-nitrate reducing, oxidase positive, catalase positive, alpha hemolytic, and citrate-positive obligate aerobe that is commonly found in the environment. A. faecalis belong to phylum Proteobacteria, class Betaproteobacteria, exploration of symbiotic and associated bacteria order Burkholderiales, and family Alcaligenaceae. IISR-EPN BC 08 and 11 shows 99% similarity A. faecalis subsp. parafaecalis strain ALK518 (accession no. KC456571) and A. faecalis subsp. parafaecalis strain ALK517 (accession no.KC456570). The optimal tree with the sum of branch length was 11.34437646. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 7 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1155 positions in the final dataset (Fig. 3 c).
Pseudomonas entomophila (IISR-EPN-BC 10)
Pseudomonas entomophila is a strain of bacterium that lives in the soil, aquatic, and rhizosphere environment soil and can infect insects. It is a gram-negative rod, facultative anaerobe, positive for catalase, oxidase, Voges-Proskauer test, and citrate utilization, whereas, it is negative for indole production, methyl red, and urease. IISR-EPNBC 10 shows 100% similarity with P. entomophila strain 2P25 (accession no. EF178450), P. entomophila strain L48 (accession no. CT573326), and P. entomophila strain L48 (accession no.NR115336). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 747 positions in the final dataset (Fig. 3 e).
Photorhabdus luminescens (IISR-EPN BC 12)
Photorhabdus is a genus of bioluminescent, gram-negative bacilli which lives symbiotically within entomopathogenic nematodes, hence the name photo (which means light-producing) and rhabdus (rod shaped) with some bipolar staining and vacuolation with occasional long filamentous forms. P. luminescens strain IISR-EPN BC 12 was facultative anaerobe, positive for catalase, indole production, citrate utilization, and methyl red, whereas it was negative for oxidase, urease, and Voges-Proskauer test. IISR-EPN BC 12 shows 100% similarity to P. luminescens subsp. akhurstii strain 0805-P5G (accession no. EU301784), P. luminescens subsp. akhurstii strain FRG04 (accession no. NR028869), and P. luminescens subsp. akhurstii strain 0813-124 (accession no. DQ223040). The optimal tree with the sum of branch length was 20.34437646. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 12 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 957 positions in the final dataset (Fig. 3 d).
This indigenous EPN Heterorhabditis sp. (IISR-EPN 09) was found to be effective against various economically important insects pests like the shoot borer Conogethes punctiferalis, hairy caterpillar Euproctis sp., cardamom root grub Basilepta fulvicorne, semi-looper Synegia sp., and leaf feeder Lema sp. (Pervez et al. 2014b, 2016, and Pervez et al. 2017).
The present study led to the identification of 5 bacterial species viz., P. luminescens, P. vermicola, A. faecalis, A. aquatilis, and P. entomophila on the basis of phenotypic, biochemical, and molecular characterizations. The nucleotide sequences of 16S rRNA genes of these isolates were submitted to GenBank and assigned accession numbers MH249776 to MH249780. Among them, P. luminescens is a well-known symbiotic bacterium and the rest are associated bacteria. Previously, associated bacteria from genera other than the well documented Photorhabdus and Xenorhabdus have been reported to be present in or on the surface of EPNs. Torres-Barragan et al. (2011) reported the presence of 4 bacterial species viz. Serratia marcescens, Enterococcus mundtii, Achromobacter xylosoxidans, Providencia rettgeri from the EPN, and Oscheius carolinensis. P. entomophila has been found to be toxic against the larvae and adults of Drosophila melanogaster and some insects belonging to 3 different orders (Vodovar et al. 2006; Ruiu 2015). For a long time, the discovery of EPN-associated bacteria more popularly referred to as “frequently associated microbiota (FAM)” has often been dismissed as laboratory/handling contaminants; but as they became frequently encountered (Walsh and Webster 2003; Gouge and Snyder 2006, and Singh et al. 2014) in the hemolymph of infected insects, researchers started paying attention to them. FAM consists predominantly of members of Proteobacteria, while other genera are also present, except for a few of them like P. entomophila, their role in killing the insect host has not been established (Ogier et al. 2020). However, their co-existence with the core symbionts even in nematodes that have been maintained for generations in laboratory conditions indicates that they may have some import roles to play, which may be discovered in future. In fact, it is now widely debated whether the pathogenicity of IJs is exclusively because of the core symbionts (Xenorhabdus and Photorhabdus) or it is a combined effect of core symbiont and FAM (Ogier et al. 2020); such studies point towards the involvement of a rather “pathobiome” and the involvement of more than one pathogen in establishing a disease as against the widely believed concept of one pathogen one disease.
Conclusion
In the present study, six bacterial isolates from the Heterorhabditis sp. (IISR-EPN 09) were identified and the latter is native to the spice plantation of the southern India. The bacteria were identified based on the phenotypic-biochemical criteria and the sequencing of 16S rRNA gene. Since the relationship between the symbiotic bacterium and the nematode is highly specific, Photorhabdus sp. from the Heterorhabditis sp., additionally some associated bacterial species belonging to genera other than Photorhabdus, were isolated and identified. These species were not reported to have a symbiotic relationship with Heterorhabditis sp. However, some associated bacteria have been found to augment the toxicity of the EPNs in some previous studies. Evaluation of the entomopathogenic potential of the symbiotic and associated bacteria against the insect pests shall be carried out to explore their toxicity potential.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ICAR:
-
Indian Council of Agricultural Research
- IISR:
-
Indian Institute of Spices Research
- EPNs:
-
Entomopathogenic nematodes
- IJs:
-
Infective juveniles
- ITS:
-
Internal transcribed spacer
- rRNA:
-
Ribosomal ribonucleic acid
- PCR:
-
Polymerase chain reaction
- BOD:
-
Biological oxygen demand
References
Akhurst RJ (1980) Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. Microbiology 121(2):303–309
Akhurst RJ, Boemare NE (1988) A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J Gen Microbiol 134(7):1835–1845
Ali SS, Ahmad R, Hussain MA, Pervez R (2005) Pest management of pulses through entomopathogenic nematodes. Indian Institute of Pulses Research, Kanpur, p 59
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Bird AF, Akhurst RJ (1983) The nature of the intestinal vesicle in nematodes of the family Steinernematidae. Int J Parasitol 13(6):599–606
David H, Kurup NK (1988) Techniques for mass production of Sturmiopsis inferens Tns. In: David H, Easwaramoorthy S (eds) Biocontrol Technology for Sugarcane Pest Management. Sugarcane Breeding Institute, Coimbatore, pp 87–92
Dillman AR, Sternberg PW (2012) Entomopathogenic nematodes. Curr Biol 22(11):430–431
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gouge DH, Snyder JL (2006) Temporal association of entomopathogenic nematodes (Rhabditida: Steinernematidae and Heterorhabditidae) and bacteria. J Invertebr Pathol 91:147–157
Kaya HK, Gaugler R (1993) Entomopathogenic nematodes. Annu Rev Entomol 38:181–206
Ogier JC, Pagès S, Frayssinet M, Gaudriault S (2020) Entomopathogenic nematodeassociated microbiota: from monoxenic paradigm to pathobiome. Microbiome 8(1):1–7
Park Y, Kim Y, Yi Y (1999) Identification and characterization of a symbiotic bacterium associated with Steinernema carpocapsae in Korea. J Asia Pacific Entomol 2:105–111
Patel JB (2001) 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol Diagn 6(4):313–321
Pervez R, Eapen SJ, Devasahayam S, Jacob TK (2014a) Natural occurrence of entomopathogenic nematodes associated with ginger (Zingiber officinale Rosc.) ecosystem in India. Indian J Nematol 42(2):238–245
Pervez R, Eapen SJ, Devasahayam S, Jacob TK (2016) Eco-friendly management of cardamom root grub (Basilepta fulvicorne Jacoby). Indian Phytopathol 69(4):260–265
Pervez R, Eapen SJ, Rajkumar (2017) Screening of native entomopathogenic nematodes against semi-looper (Synegia sp.) infesting black pepper (Piper nigrum L.). J Plant Crop 45(3):180–189
Pervez R, Jacob TK, Devasahayam S, Eapen SJ (2014b) Penetration and infectivity of entomopathogenic nematodes against Lema sp. infesting turmeric. J Spices Aromatic Crops 23(1):71–75
Pigott CR, Ellar DJ (2007) Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev 71:255–281
Ruiu L (2015) Insect pathogenic bacteria in integrated pest management. Insects 6(2):352–367
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Coldspring-harbour laboratory press, UK, p 125
Singh S, Reese JM, Casanova-Torres AM, Goodrich-Blair H, Forst S (2014) Microbial population dynamics in the hemolymph of Manduca sexta infected with Xenorhabdus nematophila and the entomopathogenic nematode Steinernema carpocapsae. Appl Environ Microbiol 80:4277–4285
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray R, Wood W, Krieg N (eds) Methods for general and molecular bacteriology. ASM Press, Washington DC, p 615
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Torres-Barragan A, Suazo A, Buhler WG, Cardoza YJ (2011) Studies on the entomopathogenicity and bacterial associates of the nematode Oscheius carolinensis. Biological Control 59(2):123–129
Vega FE, Kaya HK (2012) Insect pathology, 2nd edn. Elsevier, London, p 504
Vodovar N, Vallene D, Cruveiller S, Rouy Z, Barbe V, Acosta C, Cattolico L, Jubin C, Lajus A, Segurens B (2006) Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nature Biotechnol 24:673–679
Walsh KT, Webster JM (2003) Interaction of microbial populations in Steinernema (Steinernematidae, Nematoda) infected Galleria mellonella larvae. J Invertebr Pathol 83:118–126
Woodring JL, Kaya HK (1988) Steinernematid and heterorhabditid nematodes: a handbook of biology and techniques. Southern cooperative series bulletin 331, Arkansas Agricultural Experiment Station, Arkansas, Fayetteville 28.
Zhou J, Davey ME, Figueras JB, Rivkina E, Gilichinsky D, Tiedje JM (1997) Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913–3919
Acknowledgements
The authors are grateful to the Director, ICAR-Indian Institute of Spices Research (ICAR-IISR), Kozhikode, Kerala, India, for providing all the facilities to carry out this study. Thanks are extended to the Distributed Information Sub Centre (DISC), ICAR-Indian Institute of Spices Research, Kozhikode (Kerala), India, for their help. Thanks also to the Director, ICAR-Indian Agricultural Research Institute and Head, Division of Nematology, ICAR-Indian Agricultural Research Institute, New Delhi.
Funding
Resources from ICAR-Indian Institute of Spices Research were used for this research.
Author information
Authors and Affiliations
Contributions
RP: research concept and design, collection and/or assembly of data, data analysis and interpretation, writing the article, critical revision of the article. SAL: research concept and design, data analysis and interpretation. SP: data analysis and critical revision of the article. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pervez, R., Lone, S.A. & Pattnaik, S. Characterization of symbiotic and associated bacteria from entomopathogenic nematode Heterorhabditis sp. (nematode: Heterorhabditidae) isolated from India. Egypt J Biol Pest Control 30, 144 (2020). https://doi.org/10.1186/s41938-020-00343-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-020-00343-9