Abstract
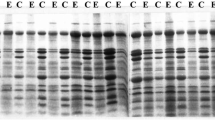
Comparative studies of two cultivated and sixteen wild species of the genus Oryza were carried out using one- and two-dimensional gel electrophoresis for variation in their seed proteins for interrelationships of these species. A number of polypeptides in the range of molecular weight 13–110 kDa were seen to vary. Under reducing conditions, polypeptides spread over the regions of mol. wt. 33–40.5, 25–27 and 19–21.5 kDa exhibited maximum variation in their patterns. Two-dimensional gel electrophoresis revealed the occurrence of disulphide-linked glutelin polypeptide pairs of mol. wt. 60, 58, 52 and 25 kDa breaking into a large and a small subunit each in the range of mol. wt. 18–40.5 and 16–25 kDa respectively in Oryza sativa. The number of such polypeptide pairs varied from 2 to 6 in different species and also in O. sativa showed variation in mol. wts. of their constituent subunits. The UPGMA dendrogram revealed that most of the Oryza species occurred in different clusters and subclusters and thus did not share very close relationships. The undisputed and closest relationship observed was that of cultivated rice O. sativa with the O. rufipogon followed by that with O. nivara. The African cultivated O. glaberrima occurring on the nearest branch of the same subcluster, thereby, supporting the phylogenetic of these species suggested in earlier studies. Eight diploid species and seven tetraploid species were included in one part of the dendrogram while the remaining two species with AA genome i.e. O. glumaepatula and O. meridionalis and one with FF i.e. O. brachyantha stood separately from these as scattered in the group of seven tetraploid species with BBCC, CCDD and HHJJ genomes. The tetraploids O. alta, O. latifolia and O. grandiglumis with CCDD genomes which occurred on the farthest part were distantly related with O. sativa. The cyanogen bromide peptide maps and two dimensional gel electrophoresis also supported the closest relationship between O. sativa and O. rufipogon.






Similar content being viewed by others
References
Aggarwal RK, Brar DS, Huang N, Khush GS (1996) Differentiation within CCDD genome species in the genus Oryza as revealed by total genomic DNA hybridization and RFLP analysis. Rice Genet Newsl 13:54–57
Aggarwal K, Brar DS, Khush GS (1997) Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Gene 254:1–12
Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS (1999) Phylogenetic relationships among Oryza species revealed by AFLP markers. Theor Appl Genet 98:1320–1328
Ali ML, Sanchez PL, Yu SB, Lorieux M, Eizenga GC (2010) Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3:218–234
Andrews AT (1990) Peptide mapping. In: Hames BD, Rickwood D (eds) Gel electrophoresis of proteins. IRL Press, Oxford, pp 301–318
Asghar R (2004) Inter and intra-specific variations in SDS-PAGE of total seed protein in rice (Oryza sativa L.) germplasm. Pak J Biol Sci 7:139–143
Bao Y, Ge S (2004) Origin and phylogeny of Oryza species with the CD genome based on multiple-gene sequence data. Plant Syst Evol 249:55–66
Bao Y, Zhou HF, Hong DY, Ge S (2006) Genetic diversity and evolutionary relationships of Oryza species with the B- and C-genomes as revealed by SSR markers. J Plant Biol 49:339–347
Brar DS, Khush GS (1997) Alien introgression in rice. Plant Mol Biol 35(1–2):35–47
Brar DS, Singh K (2011) Oryza. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, cereals. Springer, Berlin, pp 321–365
Casey R (1979) Immunoaffinity chromatography as a means of purifying legumin from Pisum (pea) seeds. Biochem J 177(2):509–520
Civáň P, Craig H, Cox CJ, Brown TA (2015) Three geographically separate domestications of Asian rice. Nat Plants 1:15164
Duan S, Lu B, Li Z, Tong J, Kong J, Yao W, Li S, Zhu Y (2007) Phylogenetic analysis of AA-genome Oryza species (Poaceae) based on chloroplast, mitochondrial, and nuclear DNA sequences. Biochem Genet 45(1–2):113–129
Federici MT, Shcherban AB, Capdevielle F, Francis M, Vaughan DA (2002) Analysis of genetic diversity in the Oryza officinalis complex. J Biotechnol 5:173–181
Galili G, Feldmen M (1984) Intergenomic suppression of endosperm protein genes in common wheat. Can J Gene Cytol 26:651–656
Ge S, Sang T, Lu BR, Hong DY (2001) Phylogeny of the genus Oryza as revealed by molecular approaches. In: Khush GS, Brar DS, Hardy B (eds) Rice genetics IV. Proceedings of the fourth international rice genetics symposium. Los Banos, The Philippines, IRRI, pp 89–105
Glazmann JC (1987) Isozymes and classification of Asian rice varieties. Theor Appl Genet 74:21–30
Hilu KW, Sharova LV (2002) Evolutionary implications of substitution pattens in prolamin genes of Oryza glaberrima (African rice, Poaceae) and related species. Am J Bot 89(2):211–219
Huang X et al (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Ishii T, Nakano T, Maeda H, Kamijima O (1996) Phylogenetic relationships in A-genome species of rice as revealed by RAPD analysis. Gene Genet Syst 71:195–201
Ishii T, Xu Y, McCouch SR (2001) Nuclear- and chloroplast-microsatellite variation in A-genome species of rice. Genome 44(4):658–666
Jaccard P (1908) Nouvelles researches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Jena KK (2010) The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice, O. sativa. Breed Sci 60:518–523
Jena KK, Kochert G (1991) Restriction fragment length polymorphism analysis of CCDD genome species of the genus Oryza L. Plant Mol Biol 16:831–839
Jiang C, Cheng Z, Zhang C, Yu T, Zhang Q, Shen JQ, Huang X (2014) Proteomic analysis of seed storage proteins in wild rice species of the Oryza genus. Prot Sci 12:51
Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao JS (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet 121:475–487
Jugran A, Bhatt ID, Rawal RS (2010) Characterization of agro-diversity by seed storage protein electrophoresis: focus on rice germplasm from Uttarakhand Himalaya. India. Rice Sci 17(2):122–128
Juliano BO (1990) Rice grain quality: problem and challenges. CFW 35(2):245–253
Kagawa H, Hirano H, Kikuchi F (1988) Variation of glutelin seed storage protein in rice. Jpn J Breed 38:327–332
Katayama T (1982) Cytogenetical studies on the genus Oryza. XIII. Relationship between the genome E and D. Jpn J Genet 57:613–621
Katsube-Tanaka T, Duldulao JB, Kimura Y, Iida S, Yamaguchi T, Nakano J, Utsumi S (2004) The two subfamilies of rice glutelin differ in both primary and higher-order structures. Biochim Biophys Acta 1699:95–102
Khan N, Katsube-Tanaka T, Iida S, Yamaguchi T, Nakano J, Tsujimoto H (2008) Identification and variation of glutelin α polypeptides in the genus Oryza assessed by two dimensional electrophoresis and step-by-step immunodetection. J Agric Food Chem 56:4955–4961
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:25–34
Krishnan HB, White AJ (1995) Morphometric analysis of rice seed protein bodies. Plant Physiol 109:1491–1495
Kumamaru T, Satoh H, Iwata N, Omura T, Omura T, Ogawa M, Tanaka K (1988) Mutants for rice storage proteins I: screening of mutants for rice storage proteins of protein bodies in the starchy endosperm. Theor Appl Genet 76:11–16
Ladizinsky G, Alder A (1976) Genetic relationships among the annual species of Cicer L. Theor Appl Genet 48:197–203
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680–688
Lee YJ, Thomson MJ, Chin JH (2014) Application of indica–japonica single-nucleotide polymorphism markers for diversity analysis of Oryza AA genome species. Plant Genet Resour 12:36–40
Lu BR (1999) Taxonomy of the genus Oryza (Poaceae): historical perspective and current status. Int Rice Res Notes 24:4–8
Lu BR, Naredo MBE, Juliano AB, Jackson MT (2000) Preliminary studies on taxonomy and biosystematics of the AA-genome. Oryza species (Poaceae). In: Jacobs SWL, Everett J (eds) Grasses, systematics & evolution. CSIRO, Melbourne, pp 51–58
Luthra R, Matta NK (1990) Seed proteins of the family Rosaceae: characterization studies and homologies. J Ind Bot Soc 69:321–324
Matta NK (1981) Structural and genetical studies on legumin of Pisum sativum L. and Vicia faba L. Ph.D. Thesis, University of Durham, UK
Matta NK, Gatehouse JA, Boulter D (1981a) The structure of legumin of Vicia faba a reappraisal. J Exp Bot 32(1):183–197
Matta NK, Gatehouse JA, Boulter D (1981b) Molecular and subunit heterogeneity of legumin of Pisum sativum (garden pea)—A multidimensional gel-electrophoretic study. J Exp Bot 32(131):1295–1307
McIntyre CL, Winberg B, Houchins K, Appels R, Baum BR (1992) Relationships between Oryza species (Poaceae) based on 5S DNA sequences. Plant Syst Evol 183:249–264
Morishima H (2001) Evolution and domestication of rice. Rice Genetics IV, IRRI, Philippines, pp 63–78
Morishima HK, Oka HI (1960) The pattern of interspecific variation in the genus Oryza: its quantitative representation by statistical methods. Evolution 14:153–165
Nair NV, Nair S, Sreenivasan TV, Mohan M (1999) Analysis of genetic diversity and phylogeny in Saccharum and related genera using RAPD markers. Genet Resour Crop Evol 46:73–79
Nayar NM (1973) Origin and cytogenetics of rice. Adv Genet 17:153–292
Ogawa M, Kumamaru T, Satoh H, Iwata N, Omura T, Kasai Z, Tanaka K (1987) Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol 28:1517–1527
Ohmido N, Fukui K (1997) Visual verification of close disposition between a rice A genome-specific DNA sequence (TrsA) and the telomere sequence. Plant Mol Biol 35:963–968
Park KC, Kim NH, Kim NS (2003) Genetic variations of AA genome Oryza species measured by MITE-AFLP. Theor Appl Genet 107:203–209
Ren FG, Lu BR, Li SQ, Huang JY, Zhu YG (2003) A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theor Appl Genet 108:113–120
Sarkar R, Bose S (1984) Electrophoretic characterization of rice varieties using single seed (salt soluble) proteins. Theor Appl Genet 68(5):415–419
Sarkar R, Raina SN (1992) Assessment of genome relationships in the genus Oryza L. based on seed-protein profile analysis. Theor Appl Genet 85:127–132
Second G (1982) Origin of the genetic diversity of cultivated rice: study of the polymorphism scored at 40 isozyme loci. Jpn J Genet 57:25–57
Second G (1985) Evolutionary relationships in the Sativa group of Oryza based on isozyme data. Genet Select Evol 17:89–114
Second G, Wang ZY (1992) Mitochondrial DNA RFLP in genus Oryza and cultivated rice. Genet Resour Crop Evol 39:125–140
Sharma SD (2003) Species of the genus Oryza and their interrelationships. In: Nanda JS, Sharma SD (eds) Monograph on the genus Oryza. Science Publishers, Enfield, pp 73–111
Siddiqui SU, Kumamaru T, Satoh H (2010) Pakistan rice genetic resources—III: SDS-PAGE diversity profile of glutelins seed storage proteins. Pak J Bot 42(4):2523–2530
Singh NP, Matta NK (2008) Variation studies on seed storage proteins and phylogenetics of the genus Cucumis. Plant Syst Evol 275:209–218
Singh NP, Matta NK (2010) Levels of seed proteins in Citrullus and Praecitrullus accessions. Plant Syst Evol 290:47–56
Singh A, Matta NK (2011) Disulphide linkages occur in many polypeptides of rice protein fractions: a two- dimensional gel electrophoretic study. Rice Sci 18:86–94
Sotowa M, Ootsuka K, Kobayashi Y, Hao Y, Tanaka K, Ichitani K, Flowers JM, Purugganan MD, Nakamura I, Sato Y (2013) Molecular relationships between Australian annual wild rice, Oryza meridionalis, and two related perennial forms. Rice 6:26
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Uppal K (1995) Studies on major seed storage proteins of Vigna mungo L. Hepper. Ph.D. Thesis. Kurukshetra University, Kurukshetra
Vaughan DA (1994) The wild relatives of rice. International Rice Research Institute, Los Banos
Vaughan DA, Morishima H (2003) Biosystematics of the genus Oryza. Chapter 1.2. In: Smith WC, Didley RH (eds) Rice. Origin, history, technology and production. Wiley, Hoboken
Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Curr Opin Plant Biol 6:139–146
Villareal RM, Juliano BO (1978) Properties of glutelin of mature and developing rice grains. Phytochemistry 17:177–182
Vithyashini L, Wickramasinghe HAM (2015) Genetic diversity of seed storage proteins of rice (Oryza sativa L.) varieties in Sri Lanka. Trop Agric Res 27(1):49–58
Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ (2015) Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci Rep. https://doi.org/10.1038/srep13957
Wang ZY, Second G, Tanksley SD (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet 83(5):565–581
Wang B, Ding Z, Liu W, Pan J, Li C, Ge S, Zhang D (2009) Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol Biol 9:250
Wang MH et al (2014) The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat Genet 46:982–988
Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, McCouch SR (1998) Identification of trait-improving quantitative trait loci alleles from wild rice relative, Oryza rufipogon. Genetics 150:899–909
Yamagata H, Sugimoto T, Tanaka K, Kasai Z (1982) Biosynthesis of storage proteins in developing rice seeds. Plant Physiol 70:1094–1100
Yin H, Akimoto M, Kaewcheenchai R, Sotowa M, Ishii T, Ishikawa R (2015) Inconsistent diversities between nuclear and plastid genomes of AA genome species in the genus Oryza. Gene Genet Syst 90:269–281
Zang LL, Zou XH, Zhang FM, Yang Z, Ge S (2011) Phylogeny and species delimitation of the C-genome diploid species in Oryza. J Syst Evol 49(5):386–395
Zhang QJ et al (2014) Rapid diversification of five Oryza AA genomes associated with rice adaptation. PNAS. https://doi.org/10.1073/pnas.1418307111
Zhu Q, Ge S (2005) Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol 167:249–265
Zou XH, Du YS, Tang L, Xu XW, Doyle JJ, Sang T, Ge S (2015) Multiple origins of BBCC allopolyploid species in the rice genus (Oryza). Sci Rep 5:14876
Acknowledgements
Kind supply of rice seeds from International Rice Research Institute, Philippines and Central Rice Research Institute (CRRI), Cuttack, India is gratefully acknowledged; AS is grateful to DAE, Trombay, India for providing financial assistance in form of Research Associateship. YK is grateful to Kurukshetra University, Kurukshetra for financial support in the form of a University Research Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Singh, A., Kumar, Y. & Matta, N.K. Electrophoretic variation in seed proteins and interrelationships of species in the genus Oryza. Genet Resour Crop Evol 65, 1915–1936 (2018). https://doi.org/10.1007/s10722-018-0665-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-018-0665-y