Summary
Ninety-three accessions representing 21 species from the genus Oryza were examined for restriction fragment length polymorphism. The majority (78%) of the accessions, for which five individuals were tested, were found to be monomorphic. Most of the polymorphic accessions segregated for only one or two probes and appeared to be mixed pure lines. For most of the Oryza species tested, the majority of the genetic variation (83%) was found between accessions from different species with only 17% between accessions within species. Tetraploid species were found to have, on average, nearly 50% more alleles (unique fragments) per individual than diploid species reflecting the allopolyploid nature of their genomes.
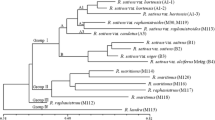
Classification of Oryza species based on RFLPs matches remarkably well previous classifications based on morphology, hybridization and isozymes. In the current study, four species complexes could be identified corresponding to those proposed by Vaughan (1989): the O. ridleyi complex, the O. meyeriana complex, the O. officinalis complex and the O. sativa complex. Within the O. sativa complex, accessions of O. rufipogon from Asia (including O. nivara) and perennial forms of O. rufipogon from Australia clustered together with accessions of cultivated rice O. sativa. Surprisingly, indica and japonica (the two major subspecies of cultivated rice) showed closer affinity with different accessions of wild O. Rufipogon than to each other, supporting a hypothesis of independent domestication events for these two types of rice. Australian annual wild rice O. meridionalis (previously classified as O. rufipogon) was clearly distinct from all other O. rufipogon accessions supporting its recent reclassification as O. meridionalis (Ng et al. 1981). Using genetic relatedness as a criterion, it was possible to identify the closest living diploid relatives of the currently known tetraploid rice species. Results from these analyses suggest that BBCC tetraploids (O. malampuzhaensis, O. punctata and O. minuta) are either of independent origins or have experienced introgression from sympatric C-genome diploid rice species. CCDD tetraploid species from America (O. latifolia, O. alta and O. grandiglumis) may be of ancient origin since they show a closer affinity to each other than to any known diploid species. Their closest living diploid relatives belong to C genome (O. eichingeri) and E genome (O. Australiensis) species. Comparisons among African, Australian and Asian rice species suggest that Oryza species in Africa and Australia are of polyphyletic origin and probably migrated to these regions at different times in the past.
Finally, on a practical note, the majority of probes used in this study detected polymorphism between cultivated rice and its wild relatives. Hence, RFLP markers and maps based on such markers are likely to be very useful in monitoring and aiding introgression of genes from wild rice into modern cultivars.
Similar content being viewed by others
References
Chang TT (1964) Present knowledge of rice genetics and cytogenetics. Tech Bull 1 Int Rice Res Inst
Chang TT, Qu SH, Pathak MD, Ling KC, Kauffman HE (1975) The search for disease and insect resistance in rice germplasm. In: Frankel OH, Hawkes JG (eds) Crop genetic resources for today and tomorrow. Cambridge University Press, Cambridge, pp 183–200
Chao S, Sharp PJ, Gale MD (1988) A linkage map of wheat homoeologous group 7 chromosomes using RFLP markers. In: Miller TE, Koebner RMD (eds) Wheat Genet Symp, vol 1. Cambridge, England, pp 493–498
Clayton WD, Renvoize SA (1986) Genera graminum. Grasses of the world. Kew Bulletin Series XIII. London, Her Majesty's Stationary Office
Coffman WR, Herrera RM (1980) Rice. In: Hybridization of crop plants. American Society of Agronomy-Crop Science of America, 28 Wisc., pp 101–131
Dally AM, Second G (1990) Chloroplast DNA diversity in wild and cultivated species of rice (genus Oryza, section Oryza). Cladistic-mutation and genetic-distance analysis. Theor Appl Genet 80:209–222
Gale MD, Sharp PJ (1988) Genetic markers in wheat — developments and prospects. In: Miller TE, Koebner RMD (eds) Proc 7th Int Wheat Genet Symp, vol 1. Cambridge, England, pp 469–475
Ghesquière A (1988) Diversité génétique de l'espèce sauvage de riz, Oryza longistaminata A. Chev. & Roehr et dynamique des flux géniques au sein du groupe Sativa. en afrique. Thèse Drès-Sciences, Université, Paris XI, Orsay
Hu Chao-Hwa (1970) Cytogenetic studies of Oryza officinalis complex. III. The genomic constitution of O. punctata and O. eichingeri. Cytologia 35:304–318
Jena KK, Khush GS (1990) Introgression of genes from Oryza officinalis Well ex WAtt to cultivated rice, O. saliva L. Theor Appl Genet 80:737–745
Katayama T (1967) Cytogenetical studies on Oryza F1 hybrids of the crosses BB x CC, BBCC x a diploid strain of O. Punctata and CC x a diploid strain of O. punctata. Proc Jpn Acad 43:327–331
Katayama T (1982) Cytogenetical studies on the genus Oryza. XIII. Relationship between the genomes E and D. Jpn J Genet 57:613–621
Kihara H (1963) Genome analysis in Oryza. In: Symp on rice problems. Tenth Pacific Sci Congr Pacific Sci Assoc, University of Hawaii, Honolulu, pp 57–66
McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829
Miller JC, Tanksley SD (1990) RFLP analysis of phylogenic relationships and genetic variation in the genus Lycopersicon. Theor Appl Genet 80:437–448
Morinaga T (1964) Cytogenetical investigation on Oryza species. In: IRRI (ed) Rice genetics and cytogenetics. Elsevier, Amsterdam, pp 91–103
Morishima H, Oka H (1960) The pattern of interspecific variation in the genus Oryza: its quantitative representation by statistical methods. Evolution 14:153–165
Morishima H, Hinata K, Oka HI (1963) Comparison of modes of evolution of cultivated forms from two wild species, Oryza breviligulata and O. perennis. Evolution 17:170–181
Morishima H, Sano Y, Oka HI (1984) Differentiation of perennial and annual types due to habitat conditions in the wild rice Oryza perennis. Plant Syst Evol 144:119–135
Nayar NM (1973) Origin and cytogenetics of rice. Advances in genetics, vol 17. Academic Press, New York London, pp 153–292
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 106–107
Ng NQ, Hawkes JG, Williams JT, Chang TT (1981) The recognition of a new species of rice (Oryza) from Australia. Bot J Linn Soc 82:327–330
Oka HI (1974) Experimental studies on the origin of cultivated rice. Genetics 78:475–486
Oka HI (1988) Origin of cultivated rice. Jpn Sci Soc Press, Amsterdam New York
Second G (1982) Origin of the genic diversity of cultivated rice (Oryza spp.): study of the polymorphism scored at 40 isozyme loci. Jpn J Genet 57:25–57
Second G (1984) Relations évolutives chez le genre Oryza et processus de domestication des riz. These Doctorat d'Etat, Université Paris-Sud, Orsay
Second G (1985a) Evolutionary relationships in the Sativa group of Oryza based on isozyme data. Genet Sel Evol 17:89–114
Second G (1985b) A new insight into the genome differentiation in Oryza L. through isozymic studies. In: Sharma AR, Sharma A (eds) Advances in chromosome and cell genetics. Oxford & IBH Pub, New Delhi Bombay Calcutta, pp 75
Sharma SD (1983) Conservation of wild species of Oryza. In: Rice germplasm conservation workshop. The International Rice Research Institute and the International Board for Plant Genetic Resources, 21–25
Sitch LA, Dalmacio RD, Romero GO (1989) Crossability of wild Oryza species and their potential use for improvement of cultivated rice. Rice Genet Lett 6:58–60
Sokal RR, Michener CD (1958) A statistic method for evaluating systematic relationships. Univ Kansas Sci Bull 28:1409–1438
Song KM, Osborn TC, William PH (1988) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs) Theor Appl Genet 75:784–794
Song KM, Osborn TC, William PH (1990) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs) 3. Genome relationships in Brassica and related genera and the origin of B. oleracea and B. rapa (syn. campestris). Theor Appl Genet 79:497–506
Tanksley SD, Young ND, Paterson AH, Bonierbale MW (1989) RFLP mapping in plant breeding — new tools for an old science. Biotechnology Bio/technology 7:257–264
Tateoka T (1962) Taxonomic studies of Oryza II. Several species complex. Bot Mag Tokyo 75:455–461
Vaughan DA (1989) The genus Oryza L. current status of taxonomy. IRRI res paper ser no 138
Vaughan DA (1990) The relatives of rice in Papua New Guinea. Report of collaborative germplasm collecting in Papua New Guinea between the Department of Primary Industry and IRRI. IRRI, Manila
Wang ZY, Tanksley SD (1989) Restriction fragment length poly-morphism in Oryza saliva L. Genome 32:1113–1118
Author information
Authors and Affiliations
Additional information
Communicated by F. Salamini
Rights and permissions
About this article
Cite this article
Wang, Z.Y., Second, G. & Tanksley, S.D. Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theoret. Appl. Genetics 83, 565–581 (1992). https://doi.org/10.1007/BF00226900
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00226900