Abstract
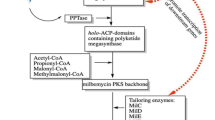
Streptomyces bingchenggensis is the main industrial producer of milbemycins, which are a group of 16-membered macrocylic lactones with excellent insecticidal activities. In the past several decades, scientists have made great efforts to solve its low productivity. However, a lack of understanding of the regulatory network of milbemycin biosynthesis limited the development of high-producing strains using a regulatory rewiring strategy. SARPs (Streptomyces Antibiotic Regulatory Proteins) family regulators are widely distributed and play key roles in regulating antibiotics production in actinobacteria. In this paper, MilR3 (encoded by sbi_06842) has been screened out for significantly affecting milbemycin production from all the 19 putative SARP family regulators in S. bingchenggensis with the DNase-deactivated Cpf1-based integrative CRISPRi system. Interestingly, milR3 is about 7 Mb away from milbemycin biosynthetic gene cluster and adjacent to a putative type II PKS (the core minimal PKS encoding genes are sbi_06843, sbi_06844, sbi_06845 and sbi_06846) gene cluster, which was proved to be responsible for producing a yellow pigment. The quantitative real-time PCR analysis proved that MilR3 positively affected the transcription of specific genes within milbemycin BGC and those from the type II PKS gene cluster. Unlike previous “small” SARP family regulators that played pathway-specific roles, MilR3 was probably a unique SARP family regulator and played a pleotropic role. MilR3 was an upper level regulator in the MilR3-MilR regulatory cascade. This study first illustrated the co-regulatory role of this unique SARP regulator. This greatly enriches our understanding of SARPs and lay a solid foundation for milbemycin yield enhancement in the near future.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Antón N, Mendes MV, Martín JF, Aparicio FJ (2004) Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J Bacteriol 186(9):2567–2575. https://doi.org/10.1128/JB.186.9.2567-2575.2004
Arias P, Fernández-Moreno MA, Malpartida F (1999) Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol 181:6958–6968. https://doi.org/10.1128/jb.181.22.6958-6968.1999
Baker GH, Blanchflower SE, Dorgen RJ, Everett JR, Manger BR, Reading CR, Readshaw SA, Shelley P (1996) Further novel milbemycin antibiotics from Streptomyces sp. E225: fermentation, isolation and structure elucidation. J Antibiot 27(35):272–280. https://doi.org/10.7164/antibiotics.49.272
Bate N, Stretigopoulos G, Gundliffe E (2002) Differential roles of two SARP-encoding regulatory genes during tylosin bisynthesis. Mol Microbiol 42(2):449–458. https://doi.org/10.1046/j.1365-2958.2002.02756.x
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8(2):208–215. https://doi.org/10.1016/j.mib.2005.02.016
Bienhoff SE, Kok DJ, Roycroft LM, Roberts ES (2013) Efficacy of a single oral administration of milbemycin oxime against natural infections of Ancylostoma braziliense in dogs. Vet Parasitol 195(1–2):102–105. https://doi.org/10.1016/j.vetpar.2013.01.004
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T (2021) antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. https://doi.org/10.1093/nar/gkab335
Carter GT, Nietsche JA, Hertz MR, William DR, Siegel MM, Morton GO, James JC, Borders DB (1988) LL-F28249 antibiotic complex: a new family of antiparastic macrocyclic lactones. J Antibiot 41(4):519–529. https://doi.org/10.7164/antibiotics.41.519
Chen Y, Wendt-Pienkowski E, Shen B (2008) Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin producntion in Streptomyces griseus ATC 49344 and heterologous hosts. J Bacteriol 190(16):5587–5596. https://doi.org/10.1128/JB.00592-08
Chen JS, Liu M, Liu XT, Miao J, Fu CZ, Gao HY, Müller R, Zhang Q, Zhang LX (2016) Interrogation of Streptomyces avermitilis for efficient production of avermectins. Synth Syst Biotechnol 1(1):7–16. https://doi.org/10.1016/j.synbio.2016.03.002
Chen S, Zhang C, Zhang L (2022) Investigation of the molecular landscape of bacterial aromatic polyketides by global analysis of type II polyketide synthases. Angew Chem Int Ed Engl 61:e202202286. https://doi.org/10.1002/anie.202202286
Danaher M, Radeck W, Kolar L, Keegan J, Cerkvenik-Flajs V (2012) Recent developments in the analysis of avermectin and milbemycin residues in food safety and the environment. Curr Pharm Biotehnol 13(6):936–995. https://doi.org/10.2174/138920112800399068
Du DY, Zhu Y, Wei JH, Tian YQ, Niu GQ, Tan HR (2013) Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene cluster. Appl Microbiol Biotechnol 97(14):6383–6396. https://doi.org/10.1007/s00253-013-4836-7
He XH, Li R, Pan YY, Liu G, Tan HR (2010a) SanG, a transcriptional activator, controls nikkomycin biosynthesis through binding to the sanN-sanO intergenic region in Streptomyces ansochromogenes. Microbiology 156(3):828–837. https://doi.org/10.1099/mic.0.033605-0
He YL, Sun YH, Liu TG, Zhou XF, Bai LQ, Deng ZX (2010b) Cloning of separate meilingmycin biosynthesis gene clusters by use of acyltransferase-ketoreductase didomain PCR amplification. Appl Environ Microbiol 76(10):3283–3292. https://doi.org/10.1128/AEM.02262-09
He HR, Ye L, Li C, Wang HY, Guo XW, Wang XJ, Zhang YY, Xiang WS (2018) SbbR/SbbA, an important ArpA/AfsA-like system, regulates milbemycin production in Streptomces bingchenggensis. Front Microbiol 5(9):1064. https://doi.org/10.3389/fmicb.2018.01064
Horinouchi S (2003) AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). J Ind Microbiol Biotechnol 30(8):462–467. https://doi.org/10.1007/s10295-003-0063-z
Jacobs CT, Scholtz CH (2015) A review on the effect of macrocyclic lactones on dung-dwelling insects: toxicity of macrocyclic lactones to dung beetles. Onderstepoort J Vet Res 82(1):858. https://doi.org/10.4102/ojvr.v82i1.858
Ji ZY, Nie QY, Yin Y, Zhang M, Pan HX, Hou XF, Tang GL (2019) Activation and characterization of cryptic gene cluster: two series of aromatic polyketides biosynthesized by divergent pathways. Angew Chem Int Ed Engl 58(50):18046–18054. https://doi.org/10.1002/anie.201910882
Jin PJ, Li SS, Zhang YY, Chu LY, He HR, Dong ZX, Xiang WS (2020) Mining and fine-tuning sugar uptake system for titer improvement of milbemycins in Streptomyces bingchenggensis. Synth Syst Biotechnol 5(3):214–221. https://doi.org/10.1016/j.synbio.2020.07.001
Kim MS, Cho WJ, Song MC, Park SW, Kim K, Kim E, Lee N, Nam SJ, Oh KH, Yoon YJ (2017) Engineered biosynthesis of milbemycins in the avermectin high-producing strain Streptomyces avermitilis. Microb Cell Fact 16(1):9. https://doi.org/10.1186/s12934-017-0626-8
Kurniawan NY, Kitani S, Maeda A, Nihira T (2014) Differential contributions of two SARP family reuglatory genes to indigoidine biosynthesis in Streptomyces lavendulae FRI-5. Appl Microbiol Biotechnol 98(23):9713–9721. https://doi.org/10.1007/s00253-014-5988-9
Lee CP, Umeyama T, Horinouchi S (2002) afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol Microbiol 43(6):1413–1430. https://doi.org/10.1046/j.1365-2958.2002.02840.x
Li R, Xie ZJ, Tian YQ, Yang HH, Chen WQ, You DL, Liu G, Deng ZX, Tan HR (2009) polR, a pathway-specific transcriptional regulatory gene, positively controls polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis. Microbiology 155(6):1819–1831. https://doi.org/10.1099/mic.0.028639-0
Li L, Wei KK, Zheng GS, Liu XC, Chen SX, Jiang WH, Lu YY (2018) CRISPR-Cpf1 assisted multiplex genome editing and transcriptional repression in Streptomyces. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00827-18
Liu G, Chater KF, Chandra G, Niu GQ, Tan HR (2013) Molecular regulation of antibiotic bisynthesis in Streptomyces. Microbiol Mol Biol Rev 77(1):112–143. https://doi.org/10.1128/MMBR.00054-12
Liu YQ, Wang HY, Li SS, Zhang YY, Cheng X, Xiang WS, Wang XJ (2021) Engineering of primary metabolic pathways for titer improvement of milbemycins in Streptomyces bingchenggensis. Appl Microbiol Biotechnol 105(5):1875–1887. https://doi.org/10.1007/s00253-021-11164-7
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Dalta C(T)) method. Method 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Ma DX, Wang C, Chen H, Wen JP (2018) Manipulating the expression of SARP family regulator BulZ and its target gene product to increase tacrolimus production. Appl Microbiol Biotechnol 102(11):4887–4900. https://doi.org/10.1007/s00253-018-8979-4
Narva KE, Feitelson JS (1990) Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J Bacteriol 172(1):326–333. https://doi.org/10.1128/jb.172.1.326-333.1990
Nicastro RL, Sato ME, Silva MZ (2011) Fitness costs associated with milbemectin resistance in the two-spotted spider mite Tetranychus urticae. Int J Pest Manag 57(3):223–228. https://doi.org/10.1080/09670874.2011.574745
Nonaka K, Kumasaka C, Okamoto Y, Maruyama F, Yoshikawa H (1999a) Bioconversion of milbemycin-related compounds: biosynthetic pathway of milbemycins. J Antibiot 52(2):109–116. https://doi.org/10.7164/antibiotics.52.109
Nonaka K, Tsukiyama T, Sato K, Kumasaka C, Maruyama F, Yoshikawa H (1999b) Bioconversion of milbemycin-related compounds: isolation and utilization of non-producer, strain RNBC-5-51. J Antibiot 52(7):620–627. https://doi.org/10.7164/antibiotics.52.620
Nonaka K, Tsukiyama T, Okamoto Y, Sato K, Kumasaka C, Yammoto T, Maruyama F, Yoshikawa H (2010) New milbemycins from Streptomyces hygroscopicus subsp. aureolacrimosus: fermantation, isolation and strucutre elucidation. J Antibiot 31(47):694–704. https://doi.org/10.7164/antibiotics.53.694
Ono M, Mishima H, Takiguchi Y, Terao M (1983) Milbemycins, a new family of macrolide antibiotics. Fermentation, isolation and physico-chemical properties of milbemycins D, E, F, G, and H. J Antibiot 36(5):509–515. https://doi.org/10.7164/antibiotics.36.509
Pluschkell U, Horowitz AR, Ishaaya I (1999) Effect of milbemectin on the sweetpotato whitefly. Bemisia Tabad Phytoparasitica 27(3):183–191. https://doi.org/10.1007/BF02981457
Romero-Rodríguez A, Robledo-Casados I, Sánchez S (2015) An overview on transcriptional regulators in Streptomyces. Biochim Biophys Acta 1894(8):1017–1039. https://doi.org/10.1016/j.bbagrm.2015.06.007
Sheldon PJ, Busarow SB, Hutchinson CR (2002) Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol Microbiol 44:449–460. https://doi.org/10.1046/j.1365-2958.2002.02886.x
Sherwood EJ, Bibb MJ (2013) The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba. Proc Natl Acad Sci U S A 110(27):E2500-2509. https://doi.org/10.1073/pnas.1305392110
Sun P, Zhao Q, Yu F, Zhang H, Wu Z, Wang Y, Wang Y, Zhang Q, Liu W (2013) Spiroketal formation and modification in avermectin biosynthesis involves a dual activity of AveC. J Am Chem Soc 135(4):1540–1548. https://doi.org/10.1021/ja311339u
Suzuki T, Mochizuki S, Yamamoto S, Arakawa K, Kinashi H (2010) Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci Biotechnol Biochem 74(4):819–827. https://doi.org/10.1271/bbb.90927
Tanaka A, Takano Y, Ohnishi Y, Horinouchi S (2007) AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Bio 369(2):322–333. https://doi.org/10.1016/j.jmb.2007.02.096
Wang XJ, Guo SL, Guo WQ, Xiang WS (2006) Role of nsdA in negative regulation of antibiotic production and morphological differentiation in Streptomyces bingchengensis. J Antibiot 62:309–313. https://doi.org/10.1038/ja.2009.33
Wang XJ, Wang XC, Xiang WS (2009) Improvement of milbemycin-producing Streptomyces bingchenggensis by rational screening of ultraviolet- and chemically induced mutants. World J Microbiol Biotechnol 25(6):1051–1056. https://doi.org/10.1007/s11274-009-9986-5
Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, Zhang J, Gao AL, Liu CX, Zhu ZX, Xiang WS (2010) Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J Bacteriol 192(17):4526–4527. https://doi.org/10.1128/JB.00596-10
Wang XJ, Zhang B, Yan YJ, An J, Zhang J, Liu CX, Xiang WS (2013) Characterization and analysis of an industrial strain of Streptomyces bingchenggensis by genome sequencing and gene microarray. Genome 56(11):677–689. https://doi.org/10.1139/gen-2013-0098
Wang HY, Zhang J, Zhang YJ, Zhang B, Liu CX, He HR, Wang XJ, Xiang WS (2014) Combined application of plasma mutagenesis and gene engineering leads to 5-oxomilbemycins A3/A4 as main components from Streptomyces bingchenggensis. Appl Microbiol Biotechnol 98(23):9703–9712. https://doi.org/10.1007/s00253-014-5970-6
Wang HY, Cheng X, Liu YQ, Li SS, Zhang YY, Wang XJ, Xiang WS (2020) Improved milbemycin production by engineering two Cytochromes P450 in Streptomyces bingchenggensis. Appl Microbiol Biotechnol 104(7):2935–2946. https://doi.org/10.1007/s00253-020-10410-8
Wei KK, Wu YJ, Li L, Jiang WH, Hu JF, Lu YH, Chen SX (2018) MilR2, a novel TetR family regulator involved in 5-oxomilbemycin A3/A4 biosynthesis in Streptomyces hygroscopicus. Appl Microbiol Biotechnol 102:8841–8853. https://doi.org/10.1007/s00253-018-9280-2
Wietzorrek A, Bibb M (1997) A novel family of proteins that regulates antibiotic production in Streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 25(6):1181–1184. https://doi.org/10.1046/j.1365-2958.1997.5421903.x
Zhang J, An J, Wang JJ, Yan YJ, He HR, Wang XJ, Xiang WS (2013) Genetic engineering of Streptomyces bingchenggensis to produce milbemycins A3/A4 as main components and eliminate the biosynthesis of nanchangmycin. Appl Microbiol Biotechnol 97(23):10091–10101. https://doi.org/10.1007/s00253-013-5255-5
Zhang YY, He HR, Liu H, Wang HY, Wang XJ, Xiang WS (2016) Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis. Microb Cell Fact 15(1):152. https://doi.org/10.1186/s12934-016-0552-1
Acknowledgements
We thank the members of Institute of Biopharmaceuticals, Taizhou University for many fruitful discussions. Many thanks to Dr. Muhammad Hafeez (Zhejiang Academy of Agricultural Sciences, Hangzhou, China) for critical editing and suggestion.
Funding
This study was funded the Natural Science Foundation of Zhejiang Province to HX (LY19C010002), the Scientific Research Foundation of Taizhou to HX (No. 2001xg07) and Taizhou University established scientific research and cultivation project (2018PY044).
Author information
Authors and Affiliations
Contributions
YY and HX: conceived and designed this study. YY, LZ, YY and LZ: performed all experimental work. HX: reviewed the results and provided critical feedback. The manuscript was drafted by YY. HX and YY: revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This manuscript is in compliance with ethical standards. This manuscript does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, YS., Yang, YQ., Zhou, LS. et al. MilR3, a unique SARP family pleiotropic regulator in Streptomyces bingchenggensis. Arch Microbiol 204, 631 (2022). https://doi.org/10.1007/s00203-022-03240-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03240-x