Abstract
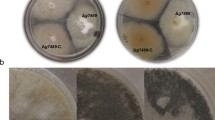
The Streptomyces antibiotic regulatory protein (SARP) family regulators have been shown to control the production of secondary metabolites in many Streptomyces species as the most downstream regulators in the regulatory cascade. Streptomyces lavendulae FRI-5 produces a blue pigment (indigoidine) together with two types of antibiotics: D-cycloserine and the nucleoside antibiotics. The production of these secondary metabolites is governed by a signaling system consisting of a γ-butyrolactone, IM-2 [(2R,3R,1′R)-2-1′-hydroxybutyl-3-hydroxymethyl-γ-butanolide], and its cognate receptor, FarA. Here, we characterized two regulatory genes of the SARP family, farR3 and farR4, which are tandemly located in the proximal region of farA. farR3 is transcribed both as a monocistronic RNA and as a bicistronic farR4-farR3 mRNA, and the expression profile is tightly controlled by the IM-2/FarA system. Loss of farR3 delayed and decreased the production of indigoidine without any changes in the transcriptional profile of other far regulatory genes, indicating that FarR3 positively controls the biosynthesis of indigoidine and is positioned in the downstream region of the IM-2/FarA signaling system. Meanwhile, loss of farR4 induced the early production of IM-2 by increasing transcription of an IM-2 biosynthetic gene, farX, indicating that FarR4 negatively controls the biosynthesis of IM-2. Thus, our results suggested differential contributions of the SARP family regulators to the regulation of secondary metabolism in S. lavendulae FRI-5. This is the first report to show that an SARP family regulator is involved in the biosynthesis of a signaling molecule functioning at the most upstream region of the regulatory cascade for Streptomyces secondary metabolism.




Similar content being viewed by others
References
Aigle B, Pang X, Decaris B, Leblond P (2005) Involvement of AlpV, a new member of the Streptomyces antibiotic regulatory protein family, in regulation of the duplicated type II polyketide synthase alp gene cluster in Streptomyces ambofaciens. J Bacteriol 187:2491–2500
Alderwick LJ, Molle V, Kremer L, Cozzone AJ, Dafforn TR, Besra GS, Fütterer K (2006) Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:2558–2563
Arias P, Fernández-moreno MA, Malpartida F (1999) Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3 (2) as a DNA-binding protein. J Bacteriol 181:6958–6968
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215
Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49
Hashimoto K, Nihira T, Sakuda S, Yamada Y (1992) IM-2, a butyrolactone autoregulator, induces production of several nucleoside antibiotics in Streptomyces sp. FRI-5. J Ferment Bioeng 73:449–455
Horinouchi S (2007) Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci Biotechnol Biochem 71:283–299
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Kitani S, Kinoshita H, Nihira T (1999) In vitro analysis of the butyrolactone autoregulator receptor protein (FarA) of Streptomyces lavendulae FRI-5 reveals that FarA acts as a DNA-binding transcriptional regulator that controls its own synthesis. J Bacteriol 181:5081–5084
Kitani S, Bibb MJ, Nihira T, Yamada Y (2000) Conjugal transfer of plasmid DNA from Escherichia coli to Streptomyces lavendulae FRI-5. J Microbiol Biotechnol 10:535–538
Kitani S, Yamada Y, Nihira T (2001) Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J Bacteriol 183:4357–4363
Kitani S, Iida A, Izumi T, Maeda A, Yamada Y, Nihira T (2008) Identification of genes involved in the butyrolactone autoregulator cascade that modulates secondary metabolism in Streptomyces lavendulae FRI-5. Gene 425:9–16
Kitani S, Doi M, Shimizu T, Maeda A, Nihira T (2010) Control of secondary metabolism by farX, which is involved in the γ-butyrolactone biosynthesis of Streptomyces lavendulae FRI-5. Arch Microbiol 192:211–220
Kuhn R, Starr MP, Kuhn DA, Bauer H, Knackmuss HJ (1965) Indigoidine and other bacterial pigments related to 3,3′-bipyridyl. Arch Microbiol 51:71–84
Liras P, Gomez-Escribano JP, Santamarta I (2008) Regulatory mechanisms controlling antibiotic production in Streptomyces clavuligerus. J Ind Microbiol Biotechnol 35:667–676
Novakova R, Odnogova Z, Kutas P, Feckova L, Kormanec J (2010) Identification and characterization of an indigoidine-like gene for a blue pigment biosynthesis in Streptomyces aureofaciens CCM 3239. Folia Microbiol 55:119–125
Novakova R, Rehakova A, Kutas P, Feckova L, Kormanec J (2011) The role of two SARP family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 157:1629–1639
Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ (1999) Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3 (2). J Bacteriol 181:204–211
Pulsawat N, Kitani S, Nihira T (2007) Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene 393:31–42
Pulsawat N, Kitani S, Fukushima E, Nihira T (2009) Hierarchical control of virginiamycin production in Streptomyces virginiae by three pathway-specific regulators: VmsS, VmsT and VmsR. Microbiology 155:1250–1259
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y (1989) Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J Ferment Bioeng 68:170–173
Sheldon PJ, Busarow SB, Hutchinson CR (2002) Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol Microbiol 44:449–460
Starr MP, Cosens G, Knackmuss HJ (1966) Formation of the blue pigment indigoidine by phytopathogenic Erwinia. Appl Microbiol 14:870–872
Takahashi H, Kumagai T, Kitani K, Mori M, Matoba Y, Sugiyama M (2007) Cloning and characterization of a Streptomyces single module type non-ribosomal peptide synthetase catalyzing a blue pigment synthesis. J Biol Chem 282:9073–9081
Takano E, Gramajo HC, Strauch E, Andres N, White J, Bibb MJ (1992) Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol Microbiol 6:2797–2804
Takano E, Nihira T, Hara Y, Jones JJ, Gershater CJL, Yamada Y, Bibb M (2000) Purification and structural determination of SCB1, a γ-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J Biol Chem 275:11010–11016
Tanaka A, Takano Y, Ohnishi Y, Horinouchi S (2007) AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369:322–333
Wang J, Wang W, Wang L, Zhang G, Fan K, Tan H, Yang K (2011) A novel role of “pseudo” γ-butyrolactone receptors in controlling γ-butyrolactone biosynthesis in Streptomyces. Mol Microbiol 82:236–250
Wietzorrek A, Bibb MJ (1997) A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an Omp-R-like DNA-binding fold. Mol Microbiol 25:1181–1184
Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H (1987) The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J Antibiot 40:496–504
Yanagimoto M, Enatsu T (1983) Regulation of a blue pigment production by γ-nonalactone in Streptomyces sp. J Ferment Technol 61:545–550
Yanagimoto M, Matsumoto K, Mori K (1988) IM2, a new inducer of blue pigment production in Streptomyces sp. MAFF 10-06015. J Ferment Technol 66:1–6
Yu Q, Du A, Liu T, Deng Z, He X (2012) The biosynthesis of the polyether antibiotic nanchangmycin is controlled by two pathway-specific transcriptional activators. Arch Microbiol 194:415–426
Acknowledgments
This work is a part of the PhD dissertation of Y.N.K. This work was supported in part by a grant for the Joint Program in the Field of Biotechnology from the Japan Society for the Promotion of Science (JSPS), the National Research Council of Thailand, and the National Science and Technology Development Agency of Thailand to T.N.; by a Grant-in-Aid for Scientific Research (B) (No. 24310157) and a Grant-in-Aid for Challenging Exploratory Research (No. 24651243) from JSPS to T.N. and S.K.; by a Grant-in-Aid for Young Scientists (B) (No. 24780071) from JSPS to S.K.; and by a scholarship from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to Y.N.K.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 407 kb)
Rights and permissions
About this article
Cite this article
Kurniawan, Y.N., Kitani, S., Maeda, A. et al. Differential contributions of two SARP family regulatory genes to indigoidine biosynthesis in Streptomyces lavendulae FRI-5. Appl Microbiol Biotechnol 98, 9713–9721 (2014). https://doi.org/10.1007/s00253-014-5988-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5988-9